Cell-free protein synthesis (CFPS) has been developed as a powerful technology and strategies to perform protein production simply and efficiently. Compared to in vivo protein production system, CFPS has several key advantages, such as rapid preparation, reduced processing timeframes, open reaction environment, the ability to use special substrates, and the ability to direct all the resources toward the production of a single product.
Prior to their evolution as a production platform, cell-free systems were originally developed as a biochemical tool to study various phenomena such as the role of translation initiation factors. The most prominent example in the history of cell-free systems came in 1961 when Marshall Nirenberg used cell-free systems to assist in his discovery of the genetic code. Over the past several decades, cell-free systems have transformed into production platforms with applications in metabolic engineering and protein synthesis. Major milestones on the protein synthesis side include the expression of human granulocyte macrophage colony stimulating factor (hGM-CSF) at the 100 L scale with titers of 700 mg/L as well as high-throughput expression of the entire human proteome.
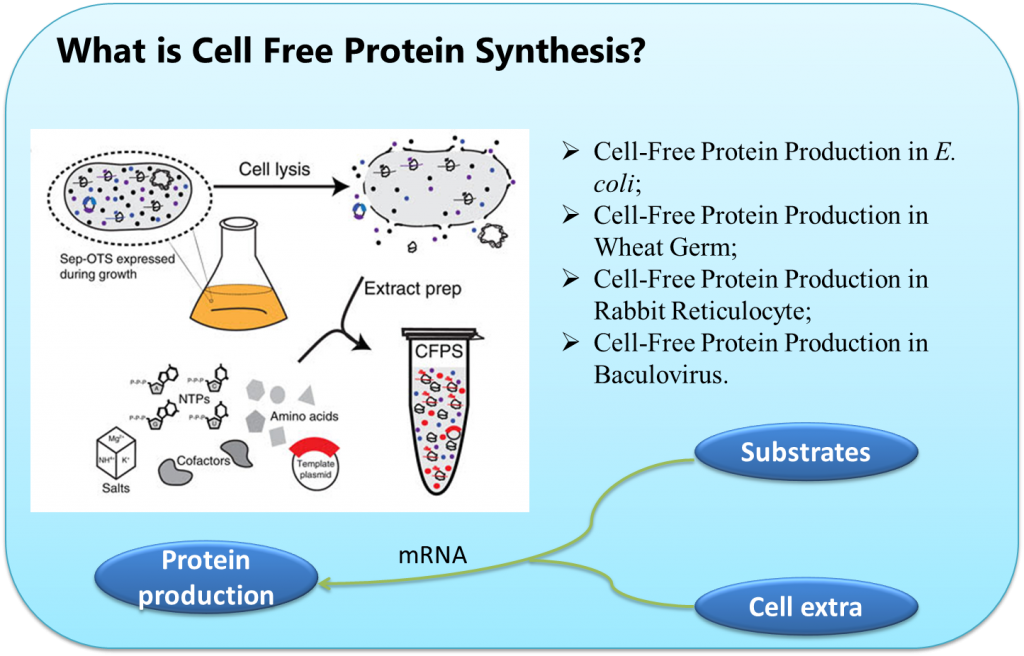
The most well developed and commonly used CFPS platform is derived from the E. coli extract. In particular, the Swartz lab at Stanford University has led the charge for developing E. coli-based CFPS by concentrating on strategies to improve yields and reduce costs. Swartz attributes the realization that central metabolism is active in the extract as the key breakthrough toward the platform’s development. This allowed for a holistic view of the platform that led to an enhanced understanding and significant improvements. Several of the many applications that can be tied to the E. coli CFPS and the Swartz lab include: the production of pharmaceutically relevant proteins such as cytokines, antibody fragments, personalized medicines and vaccine candidates, virus-like particles, and difficult to synthesize products such as [FeFe] Hydrogenases, and membrane bound proteins.
CFPS is not, however, restricted to prokaryotic-based extracts. Many eukaryotic CFPS platforms have emerged over the years including extracts based on CHO cells, HeLa cells, hybridoma cells, insect cells, protozoan Leishmania tarentola cells, and yeast cells. However, the most commonly used eukaryotic CFPS platforms to date are rabbit reticulocyte lysate and wheat germ extract. Each of these emerging eukaryotic cell-free systems have specific advantages, but these platforms are ultimately limited by their narrow appeal because they are derived from uncommon parent organism, require technically difficult extract preparation procedures that limit widespread adoption in the laboratory, or were developed strictly as a biochemical tool to investigate translation.
Powerful support for the utility of eukaryotic CFPS platforms came in 2008 when the National Institute of Advanced Industrial Science and Technology (NIAIST) in Japan used wheat germ extract to express >13,000 proteins of the human proteome. This technology utilized linear DNA templates to prime the reaction and required no cloning techniques, thus rapidly streamlining protein production timescales. This example showcases a prime example of the power of CFPS: automatable high-throughput synthesis of protein libraries.
Prior to expression of the entire human proteome, the researchers at NAIST investigated the use of both E. coli extract and wheat germ extract as their expression platform. Initially, they expressed 100 constructs using both cell-free platforms and found only 38/100 were expressed in a soluble form using E. coli extract compared to 96/100 when using wheat germ extract. This variation in soluble expression is likely due to slower translation rates of the eukaryotic ribosome and the presence of eukaryotic chaperones that can assist in folding. Although E. coli extract typically has much higher expression yields, this illustrative example showcases the strength of CFPS platforms as well as the limitations of strictly prokaryotic-based cell-free systems.
Wheat germ extract, however, is not without limitations. In particular, it relies on technically difficult processing techniques, requiring 5 kg starting material to produce 5 mL of extract, dependence on an eye selection to determine if the wheat germ is in the proper stage of growth (leading to high batch-to-batch extract variation), and up to 5 days of processing time. Furthermore, major improvements toward the development of the E. coli CFPS platform stemmed from a detailed understanding of the organism from a molecular biology perspective and ability to leverage this information to improve the CFPS platform – molecular detail that is currently lacking for wheat germ.
Thus, there remains a need to develop a robust and practical eukaryotic CFPS platform. In particular, S. cerevisiae is an excellent candidate for CFPS because, like E. coli, it can be grown quickly and under precise conditions leading to streamlined extract production and low batch-to-batch variation. Furthermore, because it is eukaryotic it is able to fold complex eukaryotic proteins and is already widely used as a biomanufacturing platform in vivo–accounting for 18.5% of all FDA and EMA licensed recombinant protein biopharmaceuticals as of January 2009. Lastly, S. cervisiae is a model organism with a vast amount of detailed information available that can be used while developing the CFPS platform.
Despite these attractive features, there has been limited research on the development of yeast-based CFPS since its origin in the 1970s and early 1980s. Instead, the majority of research involving yeast cell-free translation systems has focused on investigating translation from a fundamental perspective. In this dissertation, we reignite this research and shift the focus towards development of a robust eukaryotic CFPS platform prepared from yeast extract for bio-manufacturing applications.
Reference
- Nirenberg, M. W., Matthaei, J. H., The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci USA 1961, 47, 1588.
- Swartz, J., Developing cell-free biology for industrial applications. J Ind Microbiol Biotechnol 2006, 33, 476-485.
- Goshima, N., Kawamura, Y., Fukumoto, A., Miura, A., et al., Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat Meth 2008, 5, 1011-1017.
- Carlson E D, Gan R, Hodgman C E, et al. Cell-free protein synthesis: applications come of age[J]. Biotechnology advances, 2012, 30(5): 1185-1194.