Mempro™ Cell-Based FYVE/PHD Zinc Finger Protein Production
Creative Biostructure provides you the best services to produce high-quality FYVE/PHD zinc finger with 100% assurance because we have a team of scientific elites with years of experience and a comprehensive and advanced Mempro™ cell-based membrane protein production platform.
FYVE zinc finger is named after four proteins with zinc binding domain: Fab1, YOTB, Vac1 and EEA1. FYVE zinc finger is identified in various eukaryotic proteins related to phosphoinositide metabolism and membrane trafficking. FYVE zinc finger is a highly conserved domain about 70 residues in length, which recognizes PI3P specifically. There are three conserved sequences that define the FYVE domain: WxxD motifs located the on the N-terminal, RR/KHHCR in center, and the RVC motifs located on the C-terminal. The PHD finger is very similar to the zinc binding FYVE domain. The PHD finger is about 50-80 amino acid residues in length, and presents throughout 100 eukaryotic proteins. PHD finger generally folds as a globular protein with one alpha-helix and two beta-sheet. Some membrane proteins with PHD finger have been identified to bind to H3K4me3 while others bind to H3K9me3, mainly in association with chromatin regulation.
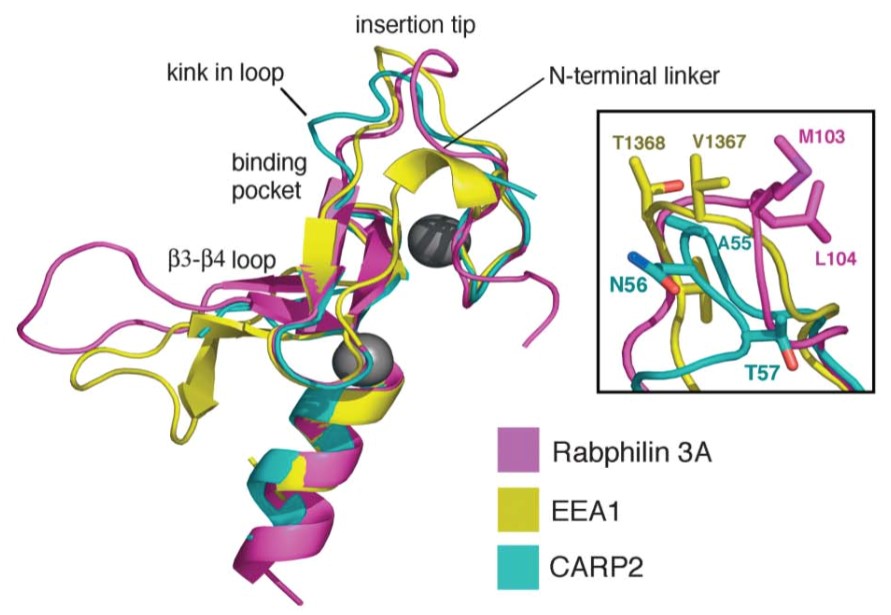 Figure 1. Unique Structural Features of the CARP2 FYVE-Type Domain. (Structure,2004)
Figure 1. Unique Structural Features of the CARP2 FYVE-Type Domain. (Structure,2004)
Creative Biostructure is focused on the high-quality and high-yield membrane proteins production services for years, we can apply various strategies for Mempro™ cell-based FYVE/PHD zinc finger production, including:
• Mempro™ FYVE/PHD Zinc Finger Protein Production in Insect Cells System
Mempro™ insect cells system has been well established by our research team for expression of membrane proteins from baculovirus-infected insect cells which is easy to scale up as well. The baculovirus expression vector system is recognized as a powerful and versatile system for high-yield membrane proteins production. Mempro™ insect cells system enables to improve solubility, perform post-translational modifications, and deliver a higher yield of target membrane protein.
• Mempro™ FYVE/PHD Zinc Finger Protein Production in Bacterial Cells System
Escherichia coli (E. coli) is the oldest but most popular used bacterial host for the membrane protein production, featuring high versatility, and providing a lot of options for the optimization of production protocols. Generally, there are two strategies can be performed in E. coli system, insertion of the soluble membrane protein into the cellular membrane, or cytoplasmic aggregation as inclusion bodies (IBs).
• Mempro™ FYVE/PHD Zinc Finger Protein Production in Mammalian Cells
Mempro™ protein production in mammalian cells leads to native membrane proteins with correct folding and post-translational modifications. We assure you that the target membrane proteins maintain the best structural and functional features.
• Mempro™ FYVE/PHD Zinc Finger Protein Production in Yeast Cells System
Single-celled yeast is an easy, quick and economic culture host, and able to apply multiple post translation modifications for eukaryotic membrane protein. Yeast cell such as saccharomyces cerevisiae and pichia pastoris, are perfect eukaryotic hosts with following advantages: high cell densities, fast growth rates, economic media, etc.
Creative Biostructure is capable for overcoming the membrane protein production difficulties by optimizing expression vectors, host strains, growth conditions, co-expression of helper proteins and fusion tags, etc, and to produce high-quality FYVE/PHD zinc finger.
We provide different strategies for Mempro™ membrane protein production services. Please feel free to contact us for a detailed quote.
References:
T. Pohlmann, et al. (2016). A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. eLife, 4:e06041.
T. G. Kutateladze, et al. (2006). Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochimica et Biophysica Acta, 1761: 868–877.
M. D. Tibbetts, et al. (2004). Crystal Structure of a FYVE-Type Zinc Finger Domain from the Caspase Regulator CARP2. Structure, 12: 2257–2263.
S. K. Elkin, et al. (2005). A PHD Finger Motif in the C Terminus of RAG2 Modulates Recombination Activity. J. Biol. Chem., 280(31): 28701–28710.