One of the most vexing tasks for biologists is to figure out how proteins, the molecules responsible for the work of cells, do their work. There are knobs, folds, and cracks on the surface of each protein, which determine what it can do. It’s fairly easy for scientists to visualize these features on a single protein. But proteins don’t act alone. Scientists also need to know the shape and composition of the complexes that proteins form when they work together.
With accurate information about the structure of protein complexes, scientists have a greater opportunity to design effective drugs to block or improve the activity of the complex, so as to achieve the purpose of treatment. They can also better predict how mutations can destroy a complex which causes disease.
But it is hard work to determine the structure of the protein complex. Each compound is different, and there is no universal way to determine its structure, and there is no way to speed up the process. Most importantly, the method to obtain the most accurate structural information requires the separation of protein complexes from their natural environment. Therefore, when scientists observe the structure, they will face a puzzling question: does it really reflect the shape of the protein complex? Does it really reflect the shape and working mechanism of protein complexes in cells?
In the final analysis, proteins come from genes. Since genes have been proved to be easier to handle than proteins, some scientists are looking for genes and a rapidly growing library of gene tools to promote the determination of protein structure.
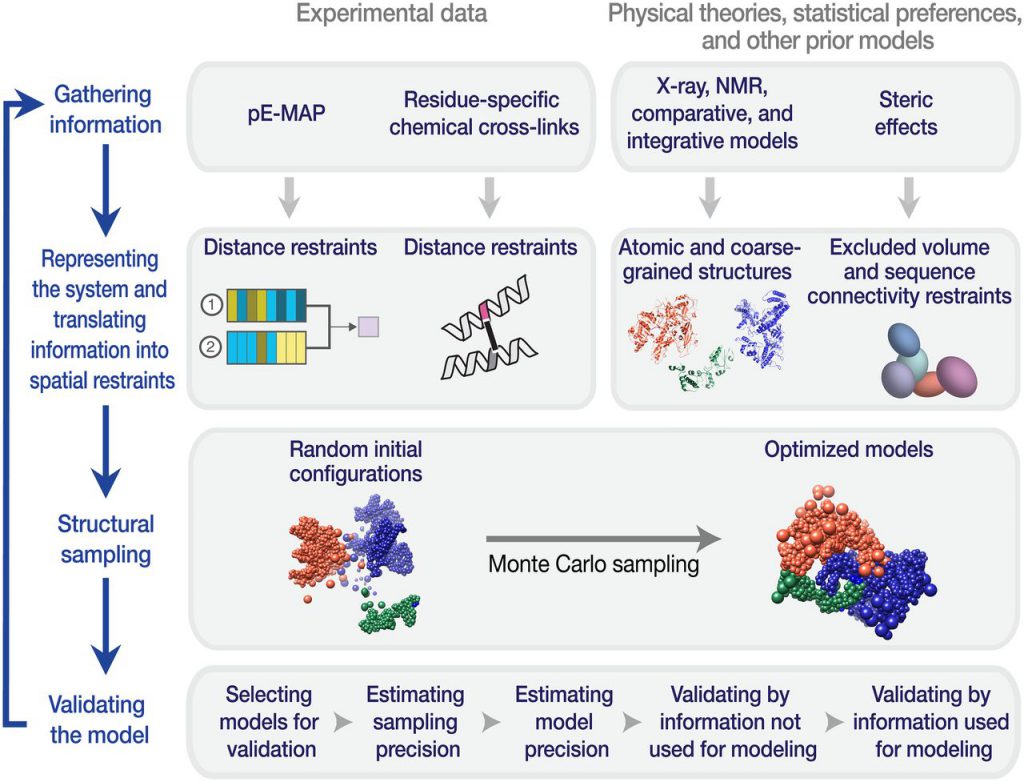
Now, in a new study, researchers from the Glaston Institute and the University of California, San Francisco have confirmed that a large-scale systematic genetic method can indeed obtain reliable and detailed structural information of protein complexes. The related research results are published in Science, with the title of “Genetic interaction mapping information integrated structure determination of protein complexes”.
Dr. Nevan Krogan, the co-author of the paper, senior researcher at Glaston Institute, and director of the quantitative bioscience research institute at the University of California, San Francisco, said, “our technology enables us to collect large amounts of structural data from living cells to reflect the way proteins work in their normal environment, rather than in artificial laboratory conditions. It should speed up the process of determining the structure of protein complexes, including those that are difficult to solve by traditional methods.”
This method is based on a technology created by Krogan called genetic interaction mapping. It can screen thousands of gene mutation combinations in living cells, and reveal the role of gene protein products in common cell processes in a relatively short time. Krogan and his team improved the resolution of these screening, and successfully identified the structural model of two protein complexes in yeast cells and one protein complex in bacterial cells.
Krogan believes that this progress is not a replacement for other methods of protein structure determination, but an important supplement. It is part of a strategy called “integrated modeling” initiated by Andrej Sali, a professor at the University of California, San Francisco.
“Combining the genetic data of the protein complexes we screened with other structural information can improve the accuracy of our model,” Krogan said, “Our research highlights the power of integrated modeling and the value of combining multiple data sets collected in completely different ways.”
Reference
- Hannes Braberg et al. Genetic interaction mapping informs integrative structure determination of protein complexes. Science, 2020, doi:10.1126/science.aaz4910.
- Dong Wang. Using genetics to reveal protein structure. Science, 2020, doi:10.1126/science.abf3863.