Professor Shi Yigong of the School of Life Sciences of Tsinghua University published the major research results on the assembly mechanism and structure of the splice body in the “Science” magazine. The paper entitled “Structures of the Fully Assembled Saccharomyces cerevisiae Spliceosome Before Activation” reports two completely assembled key conformations of the Saccharomyces cerevisiae spliceosome in the preactivated phase – Precursor pre-catalytic spliceosome (defined as “pre-B complex”) and pre-catalytic spliceosome (defined as “B complex”). The two high-resolution 3D structures with overall resolutions of 3.3-4.6 Angstroms and 3.9 Angstroms, respectively, demonstrated for the first time the recognition and dynamic status of pre-mRNA 5 ‘ splice sites and branching points (BPS) during the spliceosome assembly process. The result revealed the recognition mechanism of the 5’ splice site and BPS of the pre-mRNA before activation of the spliceosome, and how the 5’ splice site and the BPS gradually enter the active site during activation, and how the splice body gradually assembles and finally completes the activation through the structure reorganization.
RNA splicing is one of the important factors in the regulation of eukaryotic gene expression. In the 1970s, scientists discovered the discontinuity of eukaryotic genes for the first time, indicating that after genetic information transferred from DNA to RNA, it needs to undergo “shearing” and re-splicing of effective genetic information. This process of effective genetic information splicing and “invalid” genetic information removal is called RNA splicing. RNA splicing is ubiquitous in eukaryotes. With the evolution of species, the number of genes containing introns increases, and the frequency of RNA splicing increases accordingly, making it possible for one gene to encode multiple proteins. The essence of RNA splicing is a two-step transesterification reaction. This seemingly simple chemical reaction is difficult to occur on its own in cells, and there is a giant and highly dynamic molecular machine in the nucleus responsible for this chemical reaction—the spliceosome. During the splicing reaction, a variety of protein-nucleic acid complexes and splicing factors are combined and depolymerized in a highly precise sequence to form a pre-assembled complex U4/U6.U5 Tri-snRNP (U4/U6.U5 trinucleotide ribose nucleoprotein complexes) and spliceosome pre-B, B, Bact, B*, C, C*, P, and ILS complexes in at least 8 states.
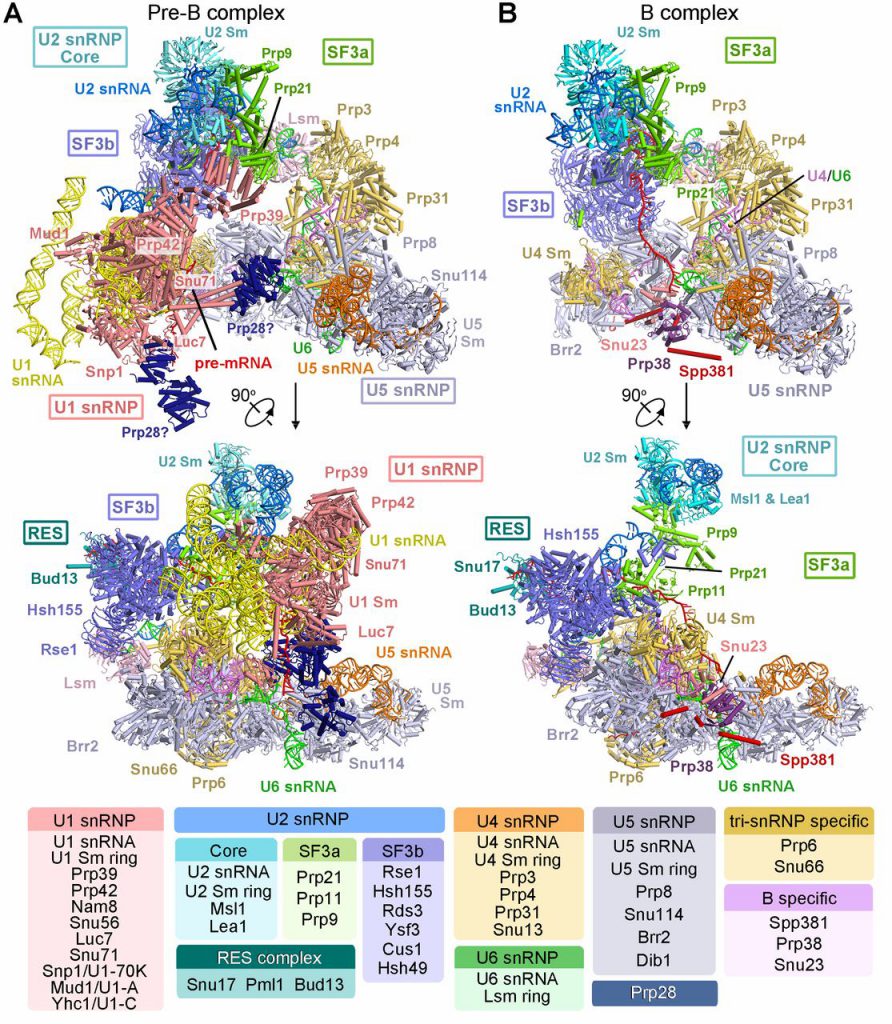
Due to the high dynamics and complexity of the splice body, the high-resolution three-dimensional structure of the spliceosome obtaining different states is recognized as a difficult problem. Under this huge challenge, Prof. Shi led the research team to overcome these difficulties. After 7 years of hard work, the high-resolution structure of 3.6 Angstroms of the fission yeast spliceosome was finally reported for the first time in 2015, revealed the catalytic center structure of nearly atomic resolution. This major research result has revolutionized the research of RNA splicing mechanism. Since the publication of the first spliceosome structure in 2015, Shi Yigong’s research group has analyzed the high-resolution structures of S. cerevisiae spliceosome complexes in six different states, which are pre-assembled complexes U4/U6. U5 Tri-snRNP of 3.8 angstroms, Bact complex at activation status of 3.5 Angstroms, first-step catalytic reaction complex C of 3.4 Angstroms, second-step catalytic activated C* complex of 4.0 Angstroms, the P complex in the post-transesterification state of 3.6 Angstroms, and the structure of the intron lasso splice construct ILS complex of 3.5 Angstroms. These resolved spliceosomes basically cover the entire RNA splicing cycle, revealing the working mechanism of the two-step reaction of spliceosome splicing RNA from the molecular level, and at the same time provide a basis for understanding the process of activation and depolymerization of the spliceosome. However, it is still difficult to explain clearly the mechanism of how the spliceosome gradually assembles and completes the activation. The two key states of the spliceosome resolved in this latest article make up for the defects in this part of the field.
The two pre-activation fully assembled spliceosome structures that were reported were challenging for every step from the purification of composites, sample preparation, to the structures analysis. The pre-B complex consists of U1 snRNP, U2 snRNP, and U4/U6.U5 tri-snRNP and is currently considered to be the largest spliceosome with the most constitutive protein. The structure of this state is complex, but the interaction between the components is not too tight, making the complex very easy to depolymerize during the purification process. In this article, Shi’s research group has repeatedly explored the purification scheme and finally optimized a set of pre-B complex samples that can be obtained with good stability. Subsequently, the structures of U1 snRNP, U2 snRNP and U4/U6.U5 tri-snRNP with the resolution of 3.3 angstroms, 3.6-4.6 angstroms and 3.4 angstroms were reconstructed using single-particle cryogenic electron microscopy.
The pre-B complex structure in this paper is the only one that has been analyzed in the world that contains five ribonucleoproteins (snRNP) spliceosome. It consists of 68 proteins and 6 RNAs. In this structure, the recognition of the 5′ splice site by U1 snRNP in the early stage of spliceosome assembly and the interaction interface between the five ribonucleoproteins were observed for the first time. At the same time, another fully assembled spliceosome structure, the high-resolution three-dimensional structure of the pre-catalyzed splice variant B complex, is reported after the pre-B complex. Combined with the structural information of B complex, through structural comparison, it can be clearly seen that the 5’ splice site of pre-mRNA was initially recognized by U1 snRNP during assembly, and was then transferred due to conformational changes and paired with U6 snRNA. This change provides the structural basis for the activation of the spliceosome. In addition, the dynamic changes of the BPS, the structural reorganization, and the components conformational changes of the splice body are clearly presented. At the end of the article, based on the structural features of pre-B, the author also boldly speculated the three-dimensional structural model of the earliest incompletely assembled pre-spliceosome (defined as “A complex”). The analysis of the structure of these two key states of the spliceosome provides the most direct and effective evidence for revealing the mechanism of how to identify the 5’ splice sites and branching points during the initial stage of spliceosome assembly, how to reconstruct the structure, and how to complete the spliceosome activation. Structural evidence will also provide structural and theoretical foundations for the study of alternative splicing of higher eukaryotes.
Reference
Rui Bai et al. Structures of the fully assembled Saccharomyces cerevisiae spliceosome before activation. Science 24 May 2018: eaau0325 DOI: 10.1126/science.aau0325