In order for viruses to proliferate, they usually need to be supported by infected cells. In many cases, the molecules they need to replicate their own genetic material are only found in the nucleus of the host cell before infecting other cells in the vicinity. But not all viruses enter the nucleus. Some viruses stay in the cytoplasm and must, therefore, be able to replicate their genetic material independently. To do so, they must bring their own “machined parts”. A key player in this process is a specialized enzyme, RNA polymerase, composed of various subunits. This enzyme reads genetic information from the viral genome and transcribes it into messenger RNA (mRNA) and uses mRNA as a blueprint for proteins encoded in the genome.
In two new studies, researchers from the BioCenter of the University of Würzburg in Germany and the Max Planck Institute for Biophysical Chemistry have now successfully solved the three-dimensional structure of vaccinia RNA polymerase (vRNAP) at atomic resolution for the first time. Vaccinia virus belongs to the poxvirus family, which is harmless to humans and forms the basis of all smallpox vaccines. Because of its favorable properties, it is currently used in the testing of oncolytic virotherapy, a new strategy against cancer. The relevant findings were recently published in the journal Cell and the papers are entitled “Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes” and “Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes,” respectively. The corresponding authors of the two papers were Dr. Utz Fischer of the University of Würzburg and Dr. Patrick Cramer of the Max Planck Institute for Biophysical Chemistry.
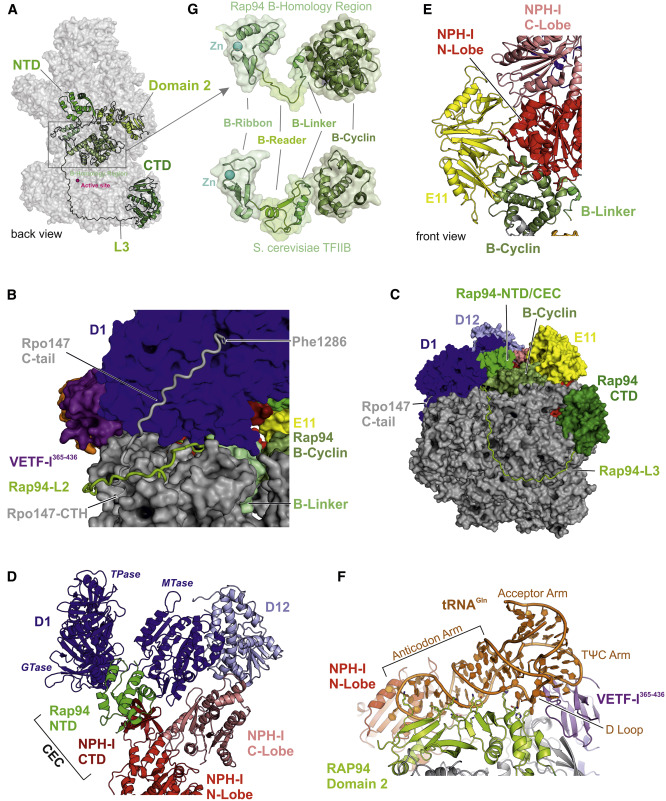
The molecular clamp that holds everything together
Fischer explained that “vRNAP exists essentially in two forms: core vRNAP and larger intact vRNAP. This complete vRNAP has various additional subunits and thus has a specialized function.” This core vRNAP is largely analogous to another known enzyme: cellular RNA polymerase II, which has also long been the focus of research in the Cramer lab. This is found in the nucleus, where it reads information on the genome and transcribes it into mRNA. Fischer refers to this complete vRNAP as the ‘all-powerful’. It consists of many subunits that can complete the entire transcription process of the virus, thus making an important step in the proliferation of this pathogen.
This complete vRNAP is held together by a molecule called transfer RNA (tRNA) that this virus lends from its host cell. Such molecules usually do not function in transcription, but provide a building block of amino acids for protein production. “Without the involvement of host tRNAs, this large molecular machine with all specific subunits would have disintegrated,” said Clements Grimm, a structural biologist at the University of Würzburg. Grimm performed the structural analysis together with Hauke Hillen at the Max Planck Institute for Biophysical Chemistry.
These researchers speculate that the host tRNA molecule performs another important task in addition to its linking function. “This tRNA is only capable of loading glutamine. Glutamine is not only an amino acid necessary for protein production, but also an important energy and nitrogen source for cells,” explains Aladar Szalay of the University of Wurzburg Cancer Treatment Research Center. The replication of this virus is nitrogen-dependent, so tRNA can act as a sensor, providing the virus with information about the current nitrogen content in the host cell. If nitrogen levels fall below a certain value, this could be a signal that the virus is leaving the host as soon as possible. But so far, this is only a guess.
To understand how this viral RNA polymerase works, these researchers also determined its three-dimensional structure at different steps of transcription. With these new findings, it is now possible to understand the entire process of virus multiplication from a structural perspective. As in the movie, it is possible to track how this molecular machine functions at the atomic level and how the individual processes are orchestrated. “It is amazing how the building blocks of this molecular machine self-rearrange to drive the synthesis of RNA products after transcription starts—this complex is indeed extremely dynamic,” explains Hillen. To gain this new insight, biochemists and structural biologists must work closely together: Biochemists Julia Bartuli and Kristina Bedenk at the University of Würzburg purified this polymerase complex with all interacting components and characterized it biochemically over the course of a year. Grimm and Hillen, structural biologists, were subsequently responsible for determining its three-dimensional structure.
Super Microscope provides the necessary data
The researchers obtained data from the latest generation of cryo-electron microscopes, devices that have revolutionized structural analysis in recent years. At a voltage of 300,000 volts, it can provide a picture at atomic resolution by emitting electrons to a sample cooled to − 180 ° C. This cryo-electron microscopy makes it possible to study biomolecules and complexes and to reconstruct their three-dimensional structures.
For about six months, Grimm and Hillen had to use computers until they obtained a spatial model of this polymerase complex from several TB of data. With three-dimensional lenses, everyone can now visualize this complex spatially, rotating it arbitrarily and breaking it down into its subunits.
Among other things, these new findings now make it possible to develop inhibitors and modulators to affect the viral proliferative cycle. Given that vaccinia virus replication occurs in the cytoplasm, these researchers also hope that it has therapeutic potential. Currently, the vaccinia virus is used for anti-cancer research in the world. Genelux has demonstrated the potential of specifically optimized vaccinia virus in shrinking tumors and detecting minimal metastases in animal experiments and patients. In addition, these researchers expect new and exciting insights into the function of related non-viral RNA polymerase complexes.
Reference
1.Clemens Grimm et al. Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes. Cell, 2019, doi:10.1016/j.cell.2019.11.024.
2.Hauke S. Hillen et al. Structural Basis of Poxvirus Transcription: Transcribing and Capping Vaccinia Complexes. Cell, 2019, doi:10.1016/j.cell.2019.11.023.
3.Virus multiplication in 3-D
https://phys.org/news/2019-12-virus-multiplication-d.html