Recently, the Xu Huaqiang group of the Shanghai Pharmaceutical Research Institute, Chinese Academy of Sciences, in cooperation with the Peter Jones group and Karsten Melcher group of the US Van Andel Research Institute, used cryo-electron microscopy to analyze the high-resolution structure of the de novo DNA methyltransferase (DNMT3A2/DNMT3B3) and the natural substrate nucleosome. Research describes the binding mode of DNMT3A2/DNMT3B3 and the nucleosome, and proposes a model of genome-wide DNA methylation. The related results are titled “Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B” and published in Nature.
DNA methylation can change the structure of chromatin, DNA stability, and the interaction between DNA and protein, thereby controlling gene expression. DNA methylation can be passed on to the new generation DNA along with the DNA replication process, which is an important epigenetic mechanism. In the chromatin environment, DNA methylation is much more complicated than in a solution. The nucleosome is the unit of genetic material, and the DNA wrapped around it is more difficult to methylate. However, most nucleosome-bound de novo DNA methyltransferases are in an inactive state. CpG methylation catalyzed by de novo DNA methyltransferases 3A and 3B is critical to mammalian development and cell differentiation, and is often closely related to the occurrence of cancer. By analyzing the expression of different subtypes of DNMT in a large number of normal tissues (GTEx database) and cancer tissues (TCGA database), this study takes the interaction of the two main DNMT subtypes DNMT3A2 and DNMT3B3 with nucleosomal nuclei in human cancer as the focus. At present, the DNMT3A catalytic domain and the DNMT3L-like catalytic domain and their crystal structures with free DNA have been resolved, but due to their limitations, the interaction mechanism between DNMT and its natural substrate nucleosome has not been elucidated.
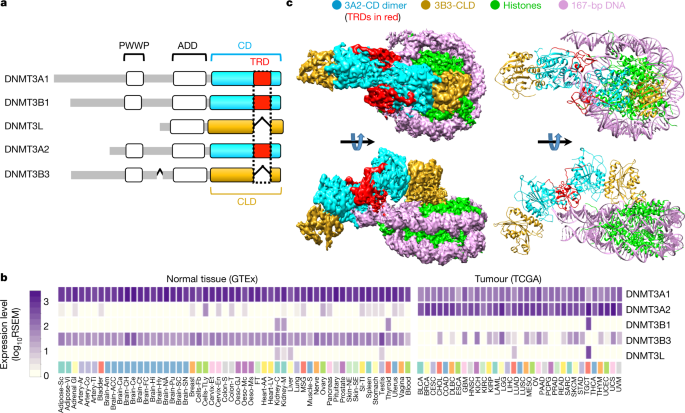
The cooperative team has long been committed to studying the important effects of DNA methylation on gene expression regulation and its extensive participation in the occurrence and development of human cancer. In order to reveal the interaction between DNMT3A2/3B3 and nucleosomes and understand DNA methylation on chromosomes, the team used cryo-electron microscopy technology to successfully analyze the structure of DNMT3A2/3B3 and nucleosome complexes with near-atomic resolution. This structure shows that the heterotetrameric complex (3B3-3A2-3A2-3B3) is very similar to the isolated DNMT3A catalytic domain and DNMT3L-type catalytic domain complex, but it interacts asymmetrically with nucleosomes. One of the DNMT3B3 catalytic domains is anchored in the acidic patch region of the nucleosome, and its core region is the arginine finger at positions 740 and 743 of DNMT3B3. The acidic patch region of nucleosomes has vital interactions with a variety of nucleosome binding proteins. However, the DNMT3A2 catalytic domain does not interact with the core region of the nucleosome, but follows the path of the DNA to interact with the linker DNA at one end and catalyze its CpG methylation. Although the DNMT3 family of proteins are highly conserved, this study revealed the switching function of the target recognition region (TRD) domain by comparing the structure of DNA-bound DNMT3A2 and the nucleosome core region-bound DNMT3B3. All catalytically active DNMT3 subtypes contain TRD domains that are critical to the target DNA. Although it sterically blocks the interaction between the catalytic domain and the core region of the nucleosome, it enhances the DNA binding ability.
In order to verify the importance of acid patch interaction for nucleosome recruitment, the study conducted mutation analysis on arginine fingers (R740 and R743), and used amino acids far away from the acid patch region (K745 and R749) as controls. In vitro interaction experiments (ALPHA Screen) showed that mutations of oppositely charged arginine fingers in the interacting core regions 740 and 743 significantly weakened the interaction between DNMT3A2/3B3 and nucleosomes, while the binding ability of non-core regions or mutations of the same charge, as expected, did not change significantly. Intracellular chromatin association assay also confirmed that mutations in the arginine fingers of 740 and 743 that interact with acid patches resulted in a decrease in chromatin binding. DNA methylation array (Infinium MethylationEPIC BeadChip) also confirmed the importance of the interaction between DNMT3B3 and nucleosome acid patches for DNA methylation reconstruction in vivo. The ability of mutant DNMT3B3 to restore DNA methylation is closely related to its ability to bind to chromatin. As a control mutation of K745 and R749, the methylation recovery level was almost the same as that of wild-type DNMT3B3. On the contrary, the R740E and R743E mutations that significantly reduce the binding of chromatin, the methylation recovery efficiency is much lower. Limited micrococcal nuclease digestion experiments (MNase digestion) further confirmed that in the presence of the DNMT complex, either side of the nucleosome significantly increased the protective region of about 10 base pairs. These data support the importance of the interaction between the catalytic domain of the DNMT complex and the acid patch for nucleosome recruitment and DNA methylation, and the binding to DNA does not depend on the active site CpG of the enzyme.
The structure and function analysis of the DNMT3A2/3B3 complex with nucleosomes revealed the unexpected nucleosome targeting function of the DNMT3B3 catalytic domain. The DNMT3A2/3B3 catalytic domain was positioned in the nucleosome connecting the DNA region, which is very important for whole-genome DNA methylation. The DNMT core nucleosome targeting and CpG methylation are separated by the DNMT-type catalytic domain and catalytic domain, which can recruit the DNMT3 complex to the vicinity of inaccessible nucleosomes, and simultaneously target CpG methylation to connect to DNA area. This suggests that the propagation of DNA methylation to nucleosomal DNA in vivo requires remodeling of the nucleosome nucleus, for example through DNA replication, transcription, or other nucleosome remodeling events.