Genomics research should not stop at obtaining information from genome sequences or epigenetic modifications. An in-depth exploration of three-dimensional chromatin folding is also essential for understanding genome functions. In the past ten years, the advancement of high-throughput sequencing technology and the development of high-resolution imaging technology have made the complex three-dimensional structure of the genome increasingly clearly presented in front of people. Among them, Topologically Associated Domains (TADs) discovered by Hi-C (high-throughput chromosome conformation capture) technology are regarded as the basic folding units for three-dimensional chromatin folding.
Nature Plants published a paper by Liu Chang’s team from the University of Tubingen, Germany, entitled “Marchantia TCP transcription factor activity correlates with three-dimensional chromatin structure”, which elaborated on the relationship between the TCP family transcription factor activity and the three-dimensional chromatin conformation changes in liverwort correlation.
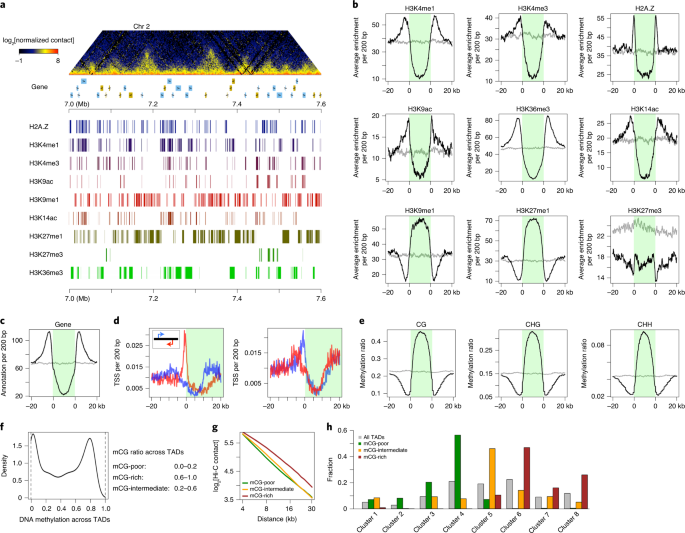
The study of animal TADs has been relatively thorough. The interior of animal TADs is a low-activity chromatin region, and its boundaries are often anchored by insulating proteins such as CTCF. TADs are involved in multiple biological processes including DNA replication and gene expression. The previous research of Liu’s group found a large number of TADs in the rice genome for the first time, and reported that the core sequences recognized by bZIP and TCP (TEOSINTE BRANCHED 1, CYCLOIDEA, AND PCF1) transcription factor families are highly enriched at the border of TADs (Nature plants: doi: 10.1038/s41477-017-0005-9, doi: 10.1038/s41477-018-0199-5). Now, TADs have been found in the genomes of many crops, but the research team has a very limited understanding of the formation and biological functions of TADs in plants.
Liu’s research team found that TADs are mainly composed of heterochromatin regions with inactive transcription and/or non-coding regions between genes. By investigating the DNA methylation level of a single TAD and the distribution of histone modifications in the TADs, they found that the performance of the liverwort TADs has the characteristics of “multi-voice harmony”: the individual TADs are different from each other, but they all have obvious epigenetic characteristics. After searching and identifying the whole genome of Dichrysanthemum morifolium, the research team found that it contains two TCP family genes TCP1 and TCP2. The two TCP genes belong to different clades. The core sequence recognized by TCP1 was significantly enriched in the border region of TADs.
ChIP-seq results show that about 11,600 regions (covering about 5% of the genome, about 11.8 Mb) on autosomes are TCP1-enriched regions (peaks), indicating that TCP1 has extensive interaction with liverwort chromatin. Among all TADs found and annotated: TCP1 peaks were detected on one side (within 1 kb) of 1164 (29%) TADs; and 499 (12%) TADs on both sides were associated with TCP1 peaks.
In addition, Liu Chang’s group identified a new type of TADs—TCP1-rich TADs. The chromatin in the TADs is loosely exposed, and about 22.6% of the MpTCP1 binding region is located in TCP1-rich TADs. Immunostaining-FISH results also showed that TCP1-rich TADs and TCP1 protein are co-localized in the nucleus in a punctate manner. TCP1-rich TADs lack methylation activation markers (such as H3K4me1, H3K4me3, and H3K36me3), and the transcriptional regulation of MpTCP1 shows the characteristics of “adapted to local conditions”, and the gene expression level in TCP1-rich TADs is relatively low. These results all indicate that the suppressive expression environment in TCP1-rich TADs is related to the TCP1 protein.
Subsequently, the team used CRISPR/Cas9 to construct a TCP1 deletion mutant. tcp1 showed the characteristics of slower growth rate and leaf curling, while the filling line can completely compensate for these growth defects. Surprisingly, there is no significant change in TADs in the Hi-C profile of tcp1 compared with Tak-1. It shows that the absence of TCP1 will not cause the edge of TADs to disappear. Although, in the tcp1 mutant, the chromatin organization pattern of border-enriched MpTCP1 TADs and TCP1-rich TADs did not change significantly. However, in the tcp1 mutant, the expression fold difference between the genes in TCP1-rich TADs and the “peripheral genes” is greater than that in the wild type.
The results of the study indicate that plant TADs, in addition to being the folding unit for the three-dimensional folding of chromatin, can also be used as “nuclear microcompartments” that regulate gene expression, providing a stable microenvironment for the genes in them.