NMR Spectroscopy Services
NMR spectroscopy is a key analytical technique for the structure elucidation of a wide range of materials from small molecules to compounds. The technique provides detailed molecular information that allows researchers to have an in-depth understanding of composition, chemical structure, morphology, and dynamics. NMR is particularly useful in the analysis of pharmaceuticals, screening weak-binding compounds and developing into drug-like inhibitors for drug discovery. Supported by our NMR platform, Creative Biostructure offers high-quality customized NMR spectroscopy services ranging from the production of labeled-proteins to acquisition and analysis of high-field NMR data for researchers in the science and pharmaceutical industry.
Advantages of Protein NMR Spectroscopy
- For proteins that are hard to obtain single crystals because of their high flexibility, NMR spectroscopy is an alternative approach to obtain high-resolution structures.
- It can measure the three-dimensional structure of biological macromolecules with atomic resolution under membrane mimetic states or almost physiological environments, especially structural analysis of low-molecular-weight proteins.
- It is an appropriate strategy for obtaining information on intermolecular interactions and biomolecular dynamics and for investigating the structure of folded intermediates and the residual structure of unfolded proteins.
- Low-transient and low-affinity complexes and small protein-ligand interactions can be studied by NMR spectroscopy.
The Workflow of Our Protein NMR Spectroscopy Services
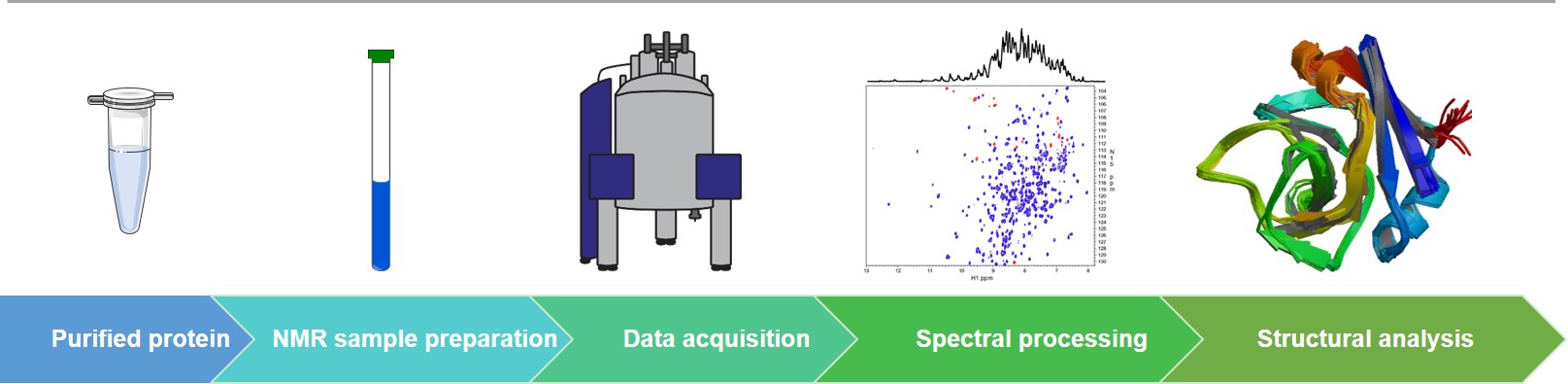
NMR Technologies at Creative Biostructure
NMR Sample Preparation Guideline
- The protein samples for structural analysis can be obtained from customers or produced by us. For larger proteins, we support the production of isotope-labeled NMR samples.
- Labeling with isotopes: Protein NMR spectroscopy requires isotope enrichment, such as 15N, 13C, 31P, 19F in common. Samples with other isotopes are also acceptable.
- Size: The molecular weight of the biomacromolecule of an NMR sample is typically less than 50 KDa for high-resolution structure elucidation. Larger proteins or complexes can be studied to identify ligand/drug-binding sites or to characterize the physical state.
- Amount: For solution NMR, a sample typically contains 1-10 mg/ml or 0.3 mM purified protein in a suitable buffer; for solid-state NMR, we require a larger amount of samples, please talk to our specialist for more information.
- Stability: For most in-depth studies by NMR, the protein needs to be stable for several days at room temperature for data acquisition.
Please feel free to contact us to discuss your project!
Ordering Process
Reference
- Xiao S, et al. Tom1 modulates binding of tollip to phosphatidylinositol 3-phosphate via a coupled folding and binding mechanism. Structure. 2015, 23(10): 1910-1920.

