Metagenomic Analysis of Coronavirus
Scientists all over the world are actively researching novel coronavirus (also known as SARS-CoV-2) and developing antiviral vaccines and drugs to cope with the global COVID-19 pandemic caused by the virus. Creative Biostructure has extensive experience in solving bioinformatics problems for genomics and proteomics. We currently provide bioinformatics solutions for viral metagenomics to support diagnostics and drug development in the field of coronavirus infection research.
Application of Viral Metagenomic Analysis in Coronavirus Research
Viral metagenomics is the study of viruses in environmental and biological samples. It utilizes next-generation sequencing (NGS) technology that generates large data sets for research. It is a pathogen detection technology that requires no cultivation and without bias. Viral metagenomics analyzes viral sequences to infer the impact of viruses on human health. Different from amplicon sequencing, metagenomics directly obtains and studies genetic material (DNA or RNA) from environmental samples, constructs metagenomic libraries, and exploits strategies of genomics research to study the genetic composition and community function of all microorganisms contained in environmental samples, and can develop new physiological active substances (or obtain novel genes). In this epidemic, researchers identified and analyzed the genome of the novel coronavirus within five days using metagenomics, which brings the time for further confirmation and prevention of the infectious pathogen and also points out for the direction for subsequent clinical diagnosis, treatment, and drug development.
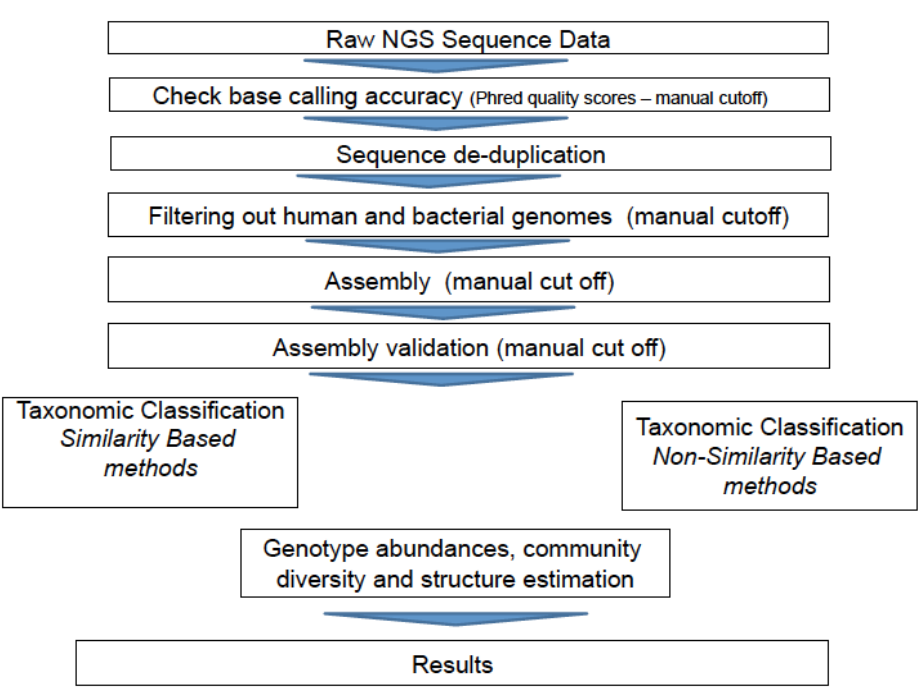 Figure 1. Bioinformatics pipeline for viral metagenomics. (Bzhalava D, Dillner J 2013)
Figure 1. Bioinformatics pipeline for viral metagenomics. (Bzhalava D, Dillner J 2013)
Our Services for Metagenomic Analysis of Coronavirus
Bioinformatics analysis is an important process of identifying and tracing new viruses using metagenomics. With sequencing technology producing millions of high-quality reads per run, processing sequence data has become a major hurdle for many researchers. At Creative Biostructure, we have a team of professional bioinformaticians with extensive experience in overcoming various challenges faced by researchers every day. With our metagenomic sequencing bioinformatics analysis service, we can provide the following information for coronavirus research:
- Analysis of species composition and abundance in respiratory or environmental samples
- Analysis of genome components
- Generation of non-redundant gene catalog and gene functional annotations
- Comparative analysis among metagenomic samples
We do not provide diagnostic tests and the service is only suitable for research purposes. If you are interested in our coronavirus-related metagenomic analysis services, please feel free to contact us. Our customer service representatives are available 24 hours a day from Monday to Sunday.
Contact us to discuss your project!
References
- Bzhalava D, Dillner J. Bioinformatics for viral metagenomics. J Data Mining Genomics Proteomics. 2013, 4(3): 2153-0602.
- Chen L.; et al. RNA based mNGS approach identifies a novel human coronavirus from two individual pneumonia cases in 2019 Wuhan outbreak. Emerging microbes & infections. 2020, 9(1): 313-319.
- Zhou P.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020: 1-4.


 Figure 1. Bioinformatics pipeline for viral metagenomics. (Bzhalava D, Dillner J 2013)
Figure 1. Bioinformatics pipeline for viral metagenomics. (Bzhalava D, Dillner J 2013)