Microscale Thermophoresis (MST) for Coronavirus Research
Microscale thermophoresis (MST) is a label-free and immobilization-free technique for quantifying biomolecular interactions. It measures the movement of molecules along a microscopic temperature gradient, which relies on changes of molecular properties in their size, charge, hydration shell or conformation, etc. By combining fluorescence detection with thermophoresis, MST provides a method to measure molecular interactions. As an advanced service provider for protein structure and function analysis, Creative Biostructure can provide MST services to help you analyze the molecular interactions involved in the coronavirus research process.
Brief Introduction to Microscale Thermophoresis (MST)
MST technique is based on thermophoresis, that is, the directional movement of molecules in a temperature gradient, to a large extent depends on various molecular characteristics, such as size, charge, hydration shell or conformation. Therefore, the technique is extremely sensitive to any changes in molecular properties, allowing accurate analysis of binding events in a few microliters of solution for almost any molecule, such as small molecules binding to proteins, ligands to liposomes or substrates to enzymes. Moreover, the MST method avoids surface immobilization and high sample consumption, giving it advantages over most commonly used non-fluorescent methods such as surface plasmon resonance (SPR) or isothermal titration calorimetry (ITC).
When performing an MST experiment, a temperature gradient is induced by an infrared laser, and the directed movement of molecules in the temperature gradient is detected and quantified using covalently linked dyes, intrinsic fluorophores, or fluorescent fusion proteins. By combining the sensitivity of thermophoresis with the accuracy of fluorescence detection, MST provides a flexible, powerful, and rapid method for the analysis of molecular interactions.
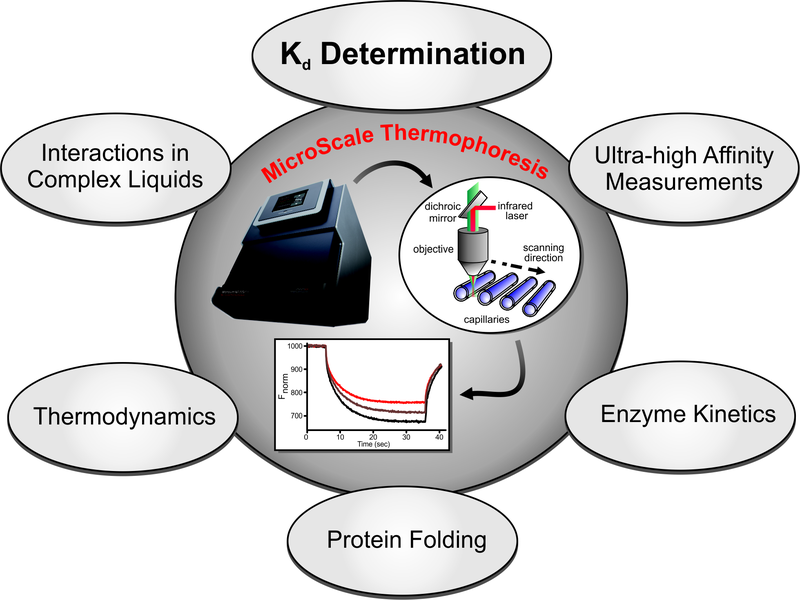
Our MST Services for Coronavirus Research
Novel coronavirus (also termed as SARS-CoV-2) is the pathogen causing the COVID-19 pandemic. Like other coronaviruses, this virus also depends on the surface Spike glycoprotein (S protein) to enter the host cell, and its molecular mechanism is mainly through the interaction between the receptor-binding domain (RBD) of S protein and ACE2. Therefore, many strategies for the development of antiviral drugs are to inhibit the entry of viruses into host cells by interfering with the binding of S protein to ACE2. The recent study utilized the MST method to identify a mini-protein capable of interacting with the RBD of SARS-CoV-2 S protein with nanomolar affinity.
Our MST services have been proven reliable by customers from academia and industry. For research on coronavirus infection, Creative Biostructure can provide you with:
- Standard biomolecular interaction studies using MST
- Determination of binding stoichiometries and binding mechanisms using MST
- Determination thermodynamic parameters for interactions using MST
- Monitoring protein folding and enzyme kinetics using MST
Our services for analysis of biomolecular interactions for coronavirus research also include surface plasmon resonance (SPR), bio-layer interferometry (BLI), isothermal titration calorimetry (ITC), fluorescence resonance energy transfer (FRET) and saturation transfer differences (STD) NMR. Want to know more? Please feel free to contact us for more information. Our customer service representatives are available 24 hours a day from Monday to Sunday.
Contact us to discuss your project!
References
- Jerabek-Willemsen M.; et al. MicroScale Thermophoresis: Interaction analysis and beyond. Journal of Molecular Structure. 2014, 1077: 101-113.
- Romano M.; et al. An engineered stable mini-protein to plug SARS-Cov2 Spikes. bioRxiv. 2020.


