Protein Evolution Analysis for Coronavirus Research
Proteins are highly adaptable and can evolve novel functions and structures. In fact, examples of the evolutionary fitness of proteins can be found everywhere, such as the numerous proteins originating from a common ancestor in nature, as well as drug resistance mechanisms caused by drug abuse, that is, the generation of various drug-resistant enzymes. Creative Biostructure supports the evolutionary analysis of coronavirus-related proteins through bioinformatics methods to help you understand the impact of local active sites on function, the overall conformational diversity of proteins, and the rate of protein evolution.
Brief Introduction to Viral Protein Evolution
Most proteins have limited stability, especially in high-pressure environments. The mutation rate of RNA virus proteins is 1 million times higher than that of bacterial or eukaryotic proteins, so the stability of RNA virus proteins is very low, which means that structures of these proteins are relatively loose, and the folding and packaging are not enough. It is easy to appear the phenomenon of local structure disorder since the tightness is not enough. However, these structural characteristics also have some advantages. For example, RNA virus proteins are highly resistant to mutations, because individual mutations have little influence on the interaction force between amino acid residues. The high degree of evolutionary ability of RNA virus proteins may also be related to their low degree of folding (i.e., insufficient interaction on the tertiary structure). Recent studies have found the existence of two mutations in the adjacent amino acid regions of non-structural protein 6 (NSP6) and open reading frame 10 (ORF 10) of novel coronavirus (SARS-CoV-2), and both of these mutations may reduce the structural stability of the protein.
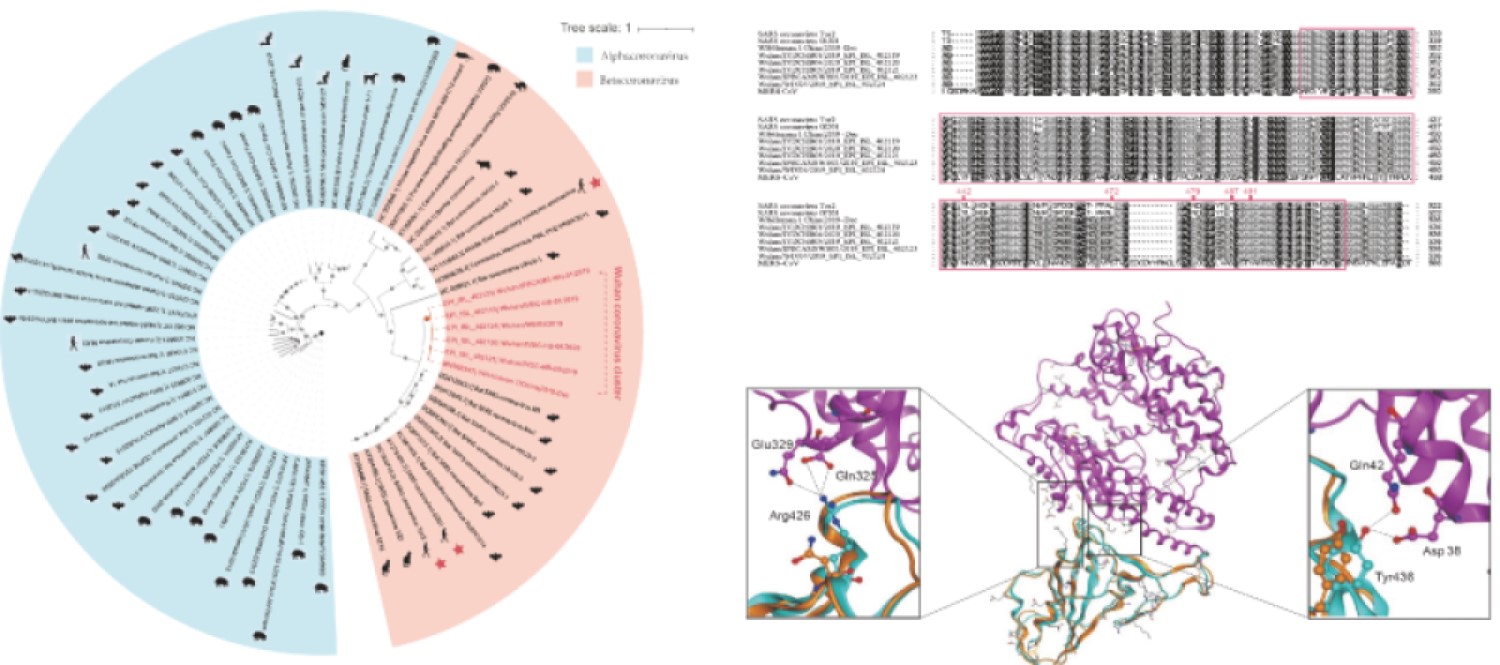 Figure 1. Evolutionary analysis of the coronaviruses and modeling of the SARS-CoV-2 Spike protein interacting with human ACE2. (Xu X.; et al. 2020)
Figure 1. Evolutionary analysis of the coronaviruses and modeling of the SARS-CoV-2 Spike protein interacting with human ACE2. (Xu X.; et al. 2020)
Protein Evolution Analysis Services for Coronavirus Research
Protein evolution analysis services for coronavirus research provided by Creative Biostructure include, but are not limited to:
- Sequence-based protein evolution analysis: Changes in protein sequence composition and the accumulation of numerous mutations help them adapt to the environment. We provide the phylogenetic tree construction service based on nucleotide and amino acid sequences to assist you in studying the rate and impact of protein evolution.
- Structure-based protein evolution analysis: In some cases of proteins with low sequence similarity preserve the fold and retain a wide range of biochemical and functional characteristics, indicating that there is an evolutionary connection. In other words, two proteins with less obvious sequence similarity may exhibit common folding and may perform similar or identical biochemical functions. We are willing to provide you with the analysis of protein evolution based on three-dimensional structures of proteins.
- Genome-based protein evolution analysis: Protein evolution is closely related to genome polymorphisms and mutations, so we believe that it is necessary to incorporate genome evolution into the protein evolution analysis.
In addition to bioinformatics methods, scientists from Creative Biostructure can also verify bioinformatics findings through experimental methods. We provide specialized mutation library construction services. Over the past few years, our site-directed mutagenesis library services have been recognized by customers from academia and industry around the world. Please feel free to contact our scientists to discuss your coronavirus-related project. Our customer service representatives are available 24 hours a day from Monday to Sunday.
Contact us to discuss your project!
References
- Tokuriki N, Tawfik D S. Protein dynamism and evolvability. Science. 2009, 324(5924): 203-207.
- Benvenuto D.; et al. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. Journal of Infection. 2020.
- Xu X.; et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Science China Life Sciences. 2020, 63(3): 457-460.

 Figure 1. Evolutionary analysis of the coronaviruses and modeling of the SARS-CoV-2 Spike protein interacting with human ACE2. (Xu X.; et al. 2020)
Figure 1. Evolutionary analysis of the coronaviruses and modeling of the SARS-CoV-2 Spike protein interacting with human ACE2. (Xu X.; et al. 2020)