Protein Structure Prediction for Coronavirus Research
Protein structure is significant to its function, so protein structure prediction is an important part of bioinformatics analysis for protein. To perform biological functions normally, protein molecules fold into one or more spatial conformations driven by many non-covalent interactions, such as hydrophobic packing, ionic interactions, hydrogen bonding, and Van der Waals forces. This is the research direction in the field of structural biology, where X-ray crystallography, NMR spectroscopy, and Cryo-EM technology are utilized to determine the three-dimensional (3D) structure of proteins. As an advanced service contract supplier in the field of structural biology, Creative Biostructure can not only provide experimental protein structure determination techniques for your coronavirus-related research but also predict the protein folding, local secondary structure and tertiary structure based merely on their amino acid sequences.
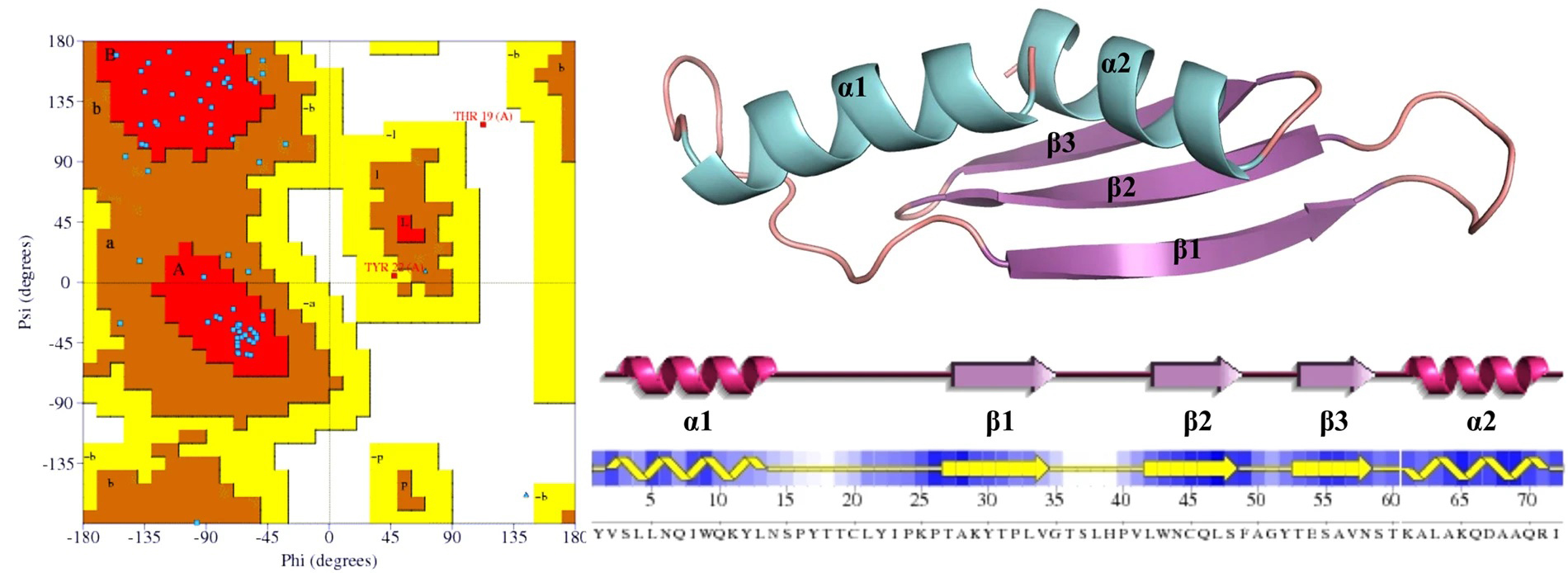 Figure 1. Structural representation of the MERS-CoV protein 4a. (Adapted from Batool M.; et al. 2017)
Figure 1. Structural representation of the MERS-CoV protein 4a. (Adapted from Batool M.; et al. 2017)
Protein Structure Prediction Services for Coronavirus Research
We exploit many bioinformatics tools for protein secondary structure prediction, protein domain structure prediction, tertiary structure prediction, and binding site analysis of proteins. The method used to predict secondary structures in proteins achieves about 80% accuracy, and we support the prediction of the secondary structure of coronavirus-related proteins using two popular neural network-based programs, PSIPRED and JPRED. In addition, we support the use of GOR program, which is more successful in predicting the α-helix than in the β-sheet. Specifically, our protein structure prediction services are as follows, including but not limited to:
- Secondary structure classification
- Non-covalent interactions analysis, such as hydrogen bond and bond analysis
- Surface analysis and side-chain analysis
- 3D structural comparison analysis
- Structure-based multiple protein sequence alignment analysis
The bioinformatics tools and approaches we support in the process of protein structure prediction include:
- Neural networks, such as PSIPRED and JPRED
- GOR method
- Chou & Fasman algorithm
- Nearest neighbor algorithm
- Hidden Markov models
- Combinatorial methods
If you are interested in our protein structure prediction services for coronavirus research, please feel free to contact us. Our customer service representatives are available 24 hours a day from Monday to Sunday, and you can contact us at any time to discuss your coronavirus-related project.
Contact us to discuss your project!
Reference
- Batool M.; et al. Structural insights into the Middle East respiratory syndrome coronavirus 4a protein and its dsRNA binding mechanism. Scientific Reports. 2017, 7(1): 1-14.

 Figure 1. Structural representation of the MERS-CoV protein 4a. (Adapted from Batool M.; et al. 2017)
Figure 1. Structural representation of the MERS-CoV protein 4a. (Adapted from Batool M.; et al. 2017)