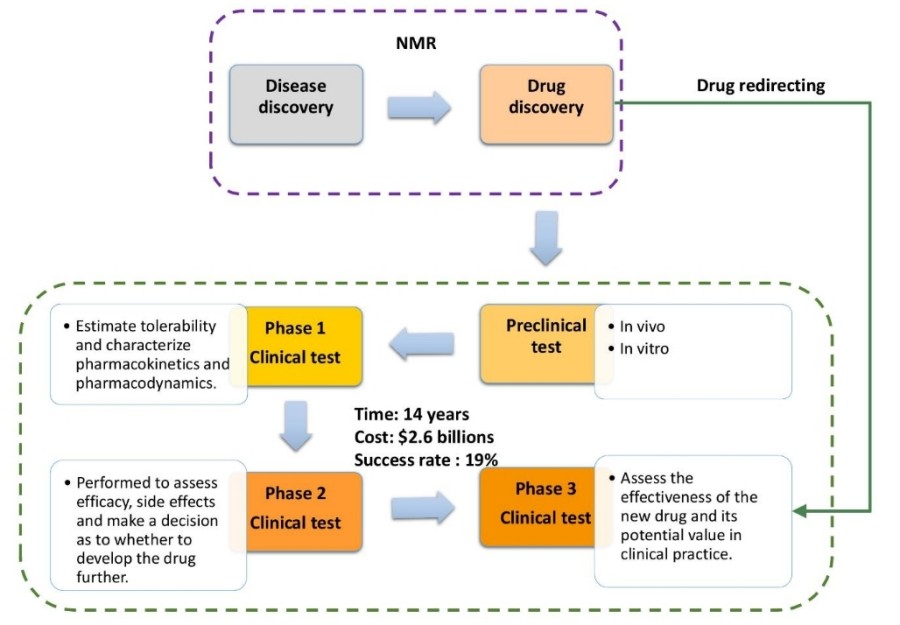
Nuclear Magnetic Resonance (NMR) spectroscopy has emerged as a pivotal tool in drug discovery, offering unique insights into molecular interactions, structural dynamics, and metabolic profiling. The pharmaceutical industry faces escalating demands to develop therapeutics efficiently. Traditional high-throughput screening (HTS) and computational methods, while effective, often overlook weak ligand-target interactions and dynamic structural details. NMR spectroscopy bridges these gaps by providing atomic-resolution data under physiological conditions, enabling fragment-based discovery, target validation, and metabolomic profiling. Explore with Creative Biostructure the multifaceted role of NMR in drug screening, highlighting its contributions to accelerating drug development.
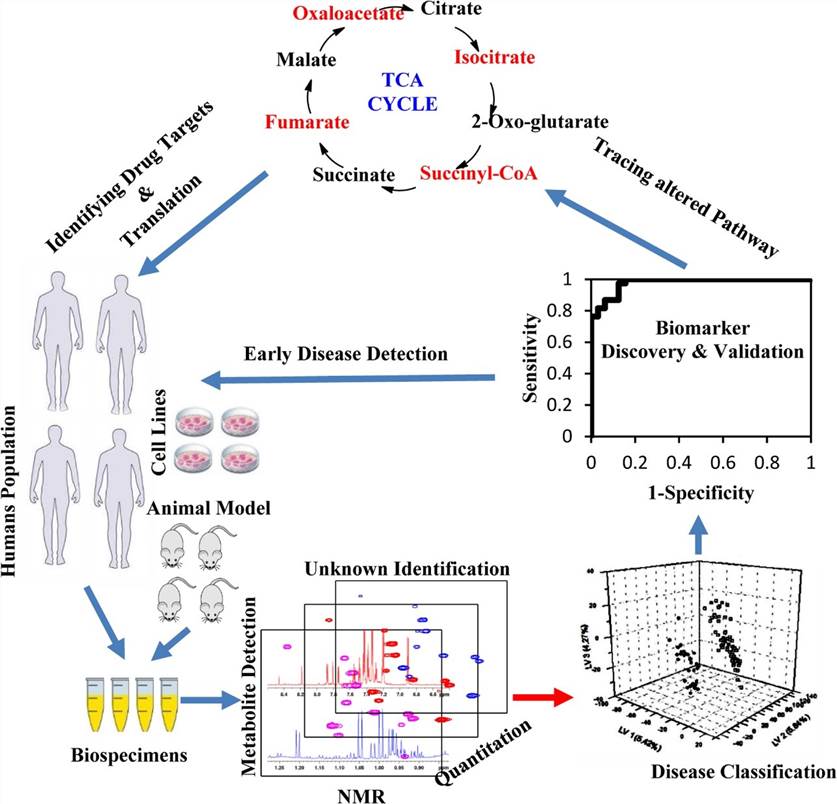
 Figure 1. Perspectives on NMR spectroscopy in drug discovery research (Caceres-Cortes et al., 2024)
Figure 1. Perspectives on NMR spectroscopy in drug discovery research (Caceres-Cortes et al., 2024)
Introduction
NMR spectroscopy plays an increasingly critical role throughout the drug discovery pipeline. Its unique ability to provide structural, dynamic, and binding information at atomic resolution, often under near-physiological conditions, makes it indispensable for early-stage screening, hit validation, lead optimization, and quality control. NMR plays a pivotal role in pharmaceutical research, supporting key applications such as fragment-based drug discovery (FBDD), target validation, and metabolomics.
Principles of NMR Relevant to Drug Screening
Basic Theory
NMR exploits the magnetic properties of nuclei (e.g., ¹H, ¹³C, ¹⁵N) in a magnetic field. When exposed to radiofrequency pulses, nuclei resonate at frequencies dependent on their chemical environment. The resulting spectra reveal structural and dynamic information through parameters like chemical shift, scalar coupling, and relaxation times.
Key Parameters
- Chemical Shift: Reflects electronic environment variations, aiding in structural elucidation.
- Relaxation Times (T1, T2): Inform on molecular mobility and binding events.
- Nuclear Overhauser Effect (NOE): Indicates spatial proximity between nuclei (<5 Å), critical for 3D structure determination.
Experimental Techniques
- 1D vs. 2D NMR: 1D for rapid analysis; 2D (e.g., NOESY, HSQC) for resolving complex mixtures.
- Isotope Filtering: Isolates signals from labeled proteins or ligands in binding studies.
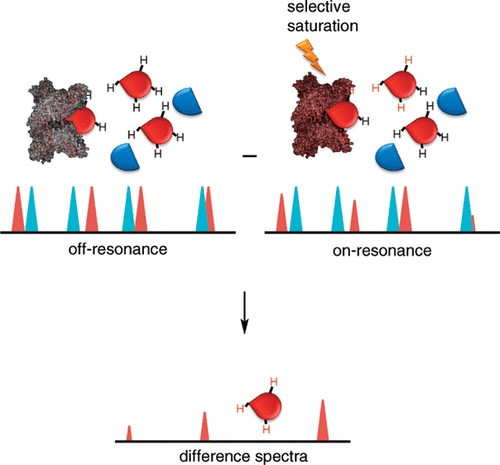
- Ligand-Observed Methods: Saturation Transfer Difference (STD) and WaterLOGSY detect ligand binding by monitoring signal attenuation.
- Target-Observed Methods: Chemical shift perturbations (CSPs) map binding sites on proteins.
Hit Identification and Fragment-Based Screening
- Fragment Libraries: Fragment-based approaches rely on screening libraries composed of small, low molecular weight (typically <300 Da) compounds that exhibit weak but specific interactions with biological targets. These fragments serve as efficient chemical starting points for the rational development of high-affinity ligands through fragment evolution, linking, or growing strategies.
- STD-NMR (Saturation Transfer Difference): STD-NMR is a ligand-detected method wherein magnetization is selectively transferred from a saturated protein to a bound ligand. This technique enables the identification of weak binders, even when the interaction is transient (Kd in the μM–mM range), making it well-suited for initial fragment screening.
- WaterLOGSY (Water-Ligand Observed via Gradient SpectroscopY): WaterLOGSY exploits the differential magnetization transfer from bulk water to free versus bound ligands. Binding events alter the phase of the NMR signal, providing a rapid and sensitive means of detecting low-affinity interactions in fragment libraries.
- Advantages of FBDD: Compared to high-throughput screening of larger, drug-like molecules, FBDD offers a higher hit rate and increased ligand efficiency. NMR-based fragment screening is particularly effective at identifying ligands for challenging targets, including protein–protein interaction interfaces, which are often refractory to conventional screening approaches.
 Figure 2. NMR applications in fragment-based drug discovery. (Shi and Zhang, 2021)
Figure 2. NMR applications in fragment-based drug discovery. (Shi and Zhang, 2021)
Hit-to-Lead Optimization
- Structure-Activity Relationship (SAR) by NMR: Pioneered by Fesik and colleagues, SAR by NMR involves sequential identification of weak-binding fragments at adjacent binding subsites, followed by their covalent or non-covalent linkage. The strategy facilitates systematic optimization of binding affinity and selectivity based on NMR-derived structural insights.
- NOE and Chemical Shift Perturbations (CSPs): The Nuclear Overhauser Effect (NOE) provides internuclear distance constraints, aiding in the elucidation of ligand orientation and protein-ligand interactions. Concurrently, CSPs in 1H-15N or 1H-13C HSQC spectra indicate perturbations in the electronic environment upon ligand binding, thereby localizing the binding site and characterizing interaction surfaces.
- Solvent Accessibility and Paramagnetic Relaxation Enhancement (PRE): PRE experiments utilize spin labels to assess the solvent exposure of ligand and protein residues. Changes in relaxation rates reveal the binding depth and conformational rearrangements, contributing to lead optimization strategies that favor deeply buried and specific interactions.
Table 1. NMR methods for hit/lead optimization. (Pellecchia et al., 2008)

Target Validation and Binding Site Characterization
- Binding Site Mapping: The combination of CSP analysis and site-directed mutagenesis allows precise mapping of ligand-binding sites, thereby validating target engagement and distinguishing between orthosteric and allosteric interactions.
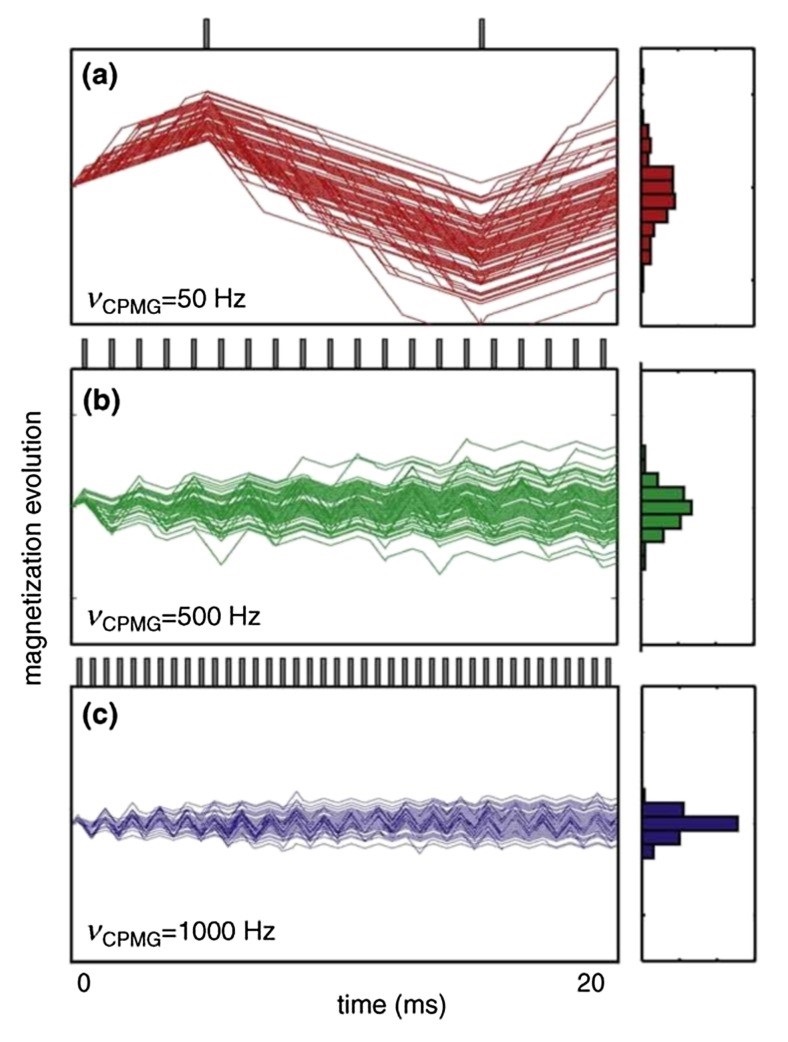
- Allosteric Site Discovery via Dynamics Measurements: NMR techniques such as Carr–Purcell–Meiboom–Gill (CPMG) relaxation dispersion and hydrogen-deuterium exchange (HDX) enable the detection of conformational dynamics associated with allosteric regulation. These methods are instrumental in identifying cryptic or transient binding pockets that may serve as novel therapeutic targets.
- Intrinsically Disordered Proteins (IDPs): NMR is uniquely suited to characterize IDPs, which lack a stable tertiary structure in the absence of a ligand. Upon ligand binding, disorder-to-order transitions can be monitored via 1H-15N HSQC and relaxation experiments, offering insights into binding mechanisms and regulatory function.
 Figure 3. Ligand-observed NMR in hit compound screening and/or validation: (a) Cartoon illustration of the Carr-Purcell-Meiboom-Gill pulse sequence (CPMG) experiment. Validation of Hsp82:SOMCL-16-171 interaction by using the CPMG technique is shown as an example. (b) Cartoon illustration of the difference spectrum in the saturation transfer difference (STD) experiment. Validation of Hsp90:1-E6 interaction by using the STD technique is shown as an example. (Shi and Zhang, 2021)
Figure 3. Ligand-observed NMR in hit compound screening and/or validation: (a) Cartoon illustration of the Carr-Purcell-Meiboom-Gill pulse sequence (CPMG) experiment. Validation of Hsp82:SOMCL-16-171 interaction by using the CPMG technique is shown as an example. (b) Cartoon illustration of the difference spectrum in the saturation transfer difference (STD) experiment. Validation of Hsp90:1-E6 interaction by using the STD technique is shown as an example. (Shi and Zhang, 2021)
Metabolomics and Toxicity Screening
- Metabolic Profiling via ¹H NMR: Proton NMR spectroscopy of biofluids (e.g., urine, plasma, serum) enables the quantification and identification of endogenous metabolites affected by drug exposure. This non-invasive approach supports the detection of biomarkers indicative of organ-specific toxicity, such as hepatotoxicity or nephrotoxicity.
- Advantages Over Mass Spectrometry (MS): While MS provides high sensitivity, NMR offers several complementary advantages: it is inherently quantitative, non-destructive, requires minimal sample preparation, and provides highly reproducible spectra. These attributes make it ideal for longitudinal toxicology studies and untargeted metabolomics.
 Figure 4. 1H-NMR-based metabolomics for cancer targeting and metabolic engineering. (Raja et al., 2020)
Figure 4. 1H-NMR-based metabolomics for cancer targeting and metabolic engineering. (Raja et al., 2020)
Quality Control and Structural Verification
- Purity and Impurity Analysis: NMR spectroscopy can accurately assess compound purity, with sensitivity to detect impurities below 2%. This capability is critical in the validation of high-throughput screening (HTS) libraries and in maintaining compound integrity during storage and handling.
- Structure Elucidation: Two-dimensional NMR experiments such as COSY, TOCSY, HSQC, and HMBC facilitate the unambiguous determination of molecular structure, stereochemistry, and connectivity. These analyses are essential for confirming the identity and consistency of synthetic compounds and reference standards.
- Regulatory Compliance: NMR is recognized by pharmacopeial standards, including the United States Pharmacopeia (USP) and the National Formulary (NF), for use in identity, purity, and potency testing. Its robustness and reproducibility ensure compliance with regulatory requirements for pharmaceutical quality assurance.
Select Service
- NMR Spectroscopy Services
- Solution-state NMR Service
- Saturation Transfer Difference (STD) NMR Service
- NMR Techniques for Structure-Based Drug Discovery
- NMR Spectroscopy in Fragment-Based Drug Design
- NMR-based Metabolomics
- CPMG Relaxation Dispersion NMR Service
- Combining NMR and Mass Spectrometry for Metabolomics
Case Studies
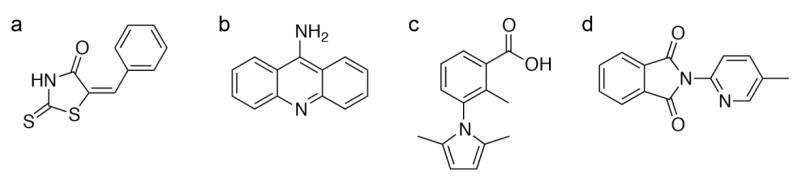
Case 1: Synthesis, stereochemistry and in vitro STD NMR and in silico HIV-1 PR enzyme-binding potential of MBH-derived inhibitors
In this study, a series of ten diastereomeric products were synthesized via aza-Michael addition reactions between a Morita–Baylis–Hillman (MBH) adduct derived from pyridine-3-carbaldehyde and various amines. The stereochemical configurations of the resulting compounds were elucidated through a combination of one-dimensional and two-dimensional NMR techniques, including NOESY, supported by computational modeling. The potential binding interactions of these compounds with HIV-1 protease subtype C were further investigated using Saturation Transfer Difference (STD) ¹H NMR spectroscopy and in silico molecular docking across five distinct HIV-1 protease receptor models.
 Figure 5. Graphic abstract for study in silico HIV-1 PR enzyme-binding potential of MBH-derived inhibitors. (Tukulula et al., 2022)
Figure 5. Graphic abstract for study in silico HIV-1 PR enzyme-binding potential of MBH-derived inhibitors. (Tukulula et al., 2022)
Case 2: 1H-NMR metabolomics reveals a multitarget action of Crithmum maritimum ethyl acetate extract in inhibiting hepatocellular carcinoma cell growth
Hepatocellular carcinoma (HCC), currently the sixth leading cause of cancer-related deaths globally. Despite available treatments, drug-related side effects remain a major concern. Plant-derived compounds have emerged as promising alternatives. This study demonstrates that the ethyl acetate extract of Crithmum maritimum, an edible plant native to Apulia, significantly inhibits HCC cell growth. Using ¹H-NMR spectroscopy, the extract was shown to disrupt multiple metabolic pathways, including suppression of the Warburg effect (via reduced lactate), inhibition of protein synthesis (via decreased amino acids), interference with membrane biosynthesis (via lower choline and phosphocholine), and modulation of lipid metabolism (reduced triglycerides, cholesterol, MUFA, DUFA, and increased PUFA). These findings suggest that Crithmum maritimum exerts a cytostatic effect through multifaceted metabolic reprogramming, supporting its potential as a preventative and therapeutic agent for HCC and possibly other cancers.
 Figure 6. 1H-NMR typical spectrum (600 MHz, CD3OD:CDCl3) of Crithmum maritimum ethyl acetate extract sample in (a) high, (b) middle, and (c) low-frequency regions, showing the main metabolite classes associated with principal peaks. (Gnocchi et al., 2021)
Figure 6. 1H-NMR typical spectrum (600 MHz, CD3OD:CDCl3) of Crithmum maritimum ethyl acetate extract sample in (a) high, (b) middle, and (c) low-frequency regions, showing the main metabolite classes associated with principal peaks. (Gnocchi et al., 2021)
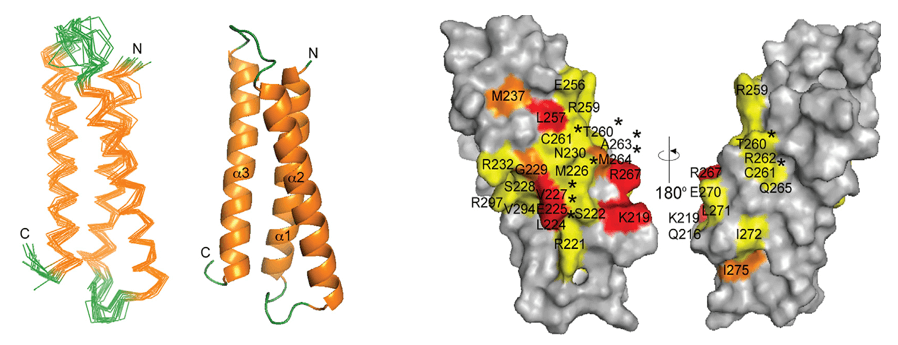
Case 3: NMR three-dimensional structure of the cationic peptide Stigmurin from Tityus stigmurus scorpion venom: In vitro antioxidant and in vivo antibacterial and healing activity
The rise of antimicrobial resistance underscores the urgent need for new antibacterial agents. In this context, Stigmurin, a 17-residue linear peptide derived from the venom of the scorpion Tityus stigmurus, emerges as a promising candidate. Notably, this study reports—for the first time—the three-dimensional structure of a disulfide-free venom peptide from the Tityus genus, determined via high-resolution NMR spectroscopy. The NMR analysis revealed a flexible N-terminal segment (Phe1–Pro6) and a defined α-helical region spanning Ser7–Phe16 in a trifluoroethanol:water environment. The combination of NMR-defined structural data and multifaceted biological activity highlights Stigmurin as a strong scaffold for the rational design of novel antimicrobial therapeutics targeting resistant bacterial strains.
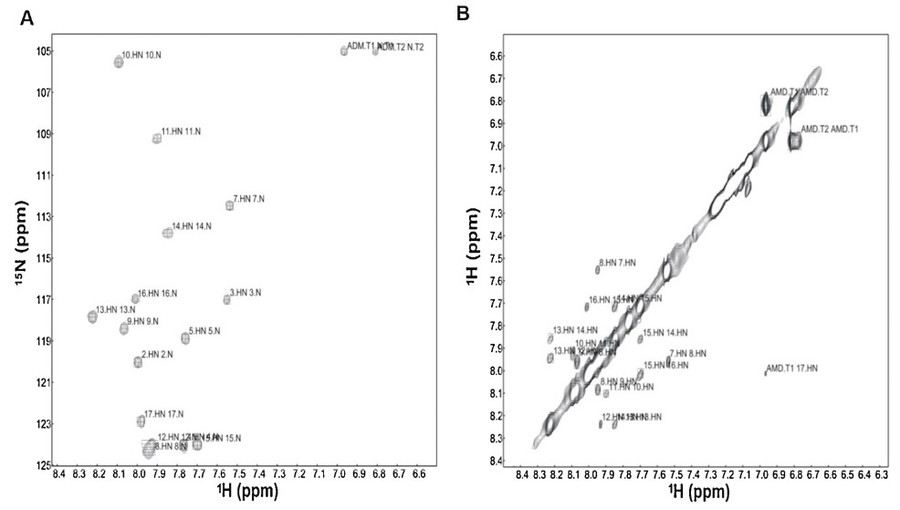 Figure 7. (A) Expansion of the 1H-15N-HMQC and (B) NH-NH correlations contour map of 5.0 mM Stigmurin in TFE-d2:H2O (40:60, v:v), 298 K. (Daniele-Silva et al., 2020)
Figure 7. (A) Expansion of the 1H-15N-HMQC and (B) NH-NH correlations contour map of 5.0 mM Stigmurin in TFE-d2:H2O (40:60, v:v), 298 K. (Daniele-Silva et al., 2020)
NMR spectroscopy offers unmatched precision in early-stage drug screening, enabling researchers to identify promising leads with confidence and efficiency. Ready to improve your screening strategy? Contact us today to learn how our NMR expertise can support your drug development pipeline.
References
- Caceres-Cortes J, Falk B, Mueller L, Dhar TGM. Perspectives on nuclear magnetic resonance spectroscopy in drug discovery research. J Med Chem. 2024;67(3):1701-1733. https://doi.org/10.1021/acs.jmedchem.3c02389
- Daniele-Silva A, Rodrigues SDCS, Dos Santos ECG, et al. NMR three-dimensional structure of the cationic peptide Stigmurin from Tityus stigmurus scorpion venom: In vitro antioxidant and in vivo antibacterial and healing activity. Peptides. 2021;137:170478. https://doi.org/10.1016/j.peptides.2020.170478
- Gnocchi D, Del Coco L, Girelli CR, et al. 1H-NMR metabolomics reveals a multitarget action of Crithmum maritimum ethyl acetate extract in inhibiting hepatocellular carcinoma cell growth. Sci Rep. 2021;11(1):1259. https://doi.org/10.1038/s41598-020-78867-1
- Gossert AD, Jahnke W. NMR in drug discovery: A practical guide to identification and validation of ligands interacting with biological macromolecules. Progress in Nuclear Magnetic Resonance Spectroscopy. 2016;97:82-125. https://doi.org/10.1016/j.pnmrs.2016.09.001
- Moore JM. NMR screening in drug discovery. Current Opinion in Biotechnology. 1999;10(1):54-58. https://doi.org/10.1016/S0958-1669(99)80010-X
- Pellecchia M, Bertini I, Cowburn D, et al. Perspectives on NMR in drug discovery: a technique comes of age. Nat Rev Drug Discov. 2008;7(9):738-745. https://doi.org/10.1038/nrd2606
- Raja G, Jung Y, Jung SH, Kim TJ. 1H-NMR-based metabolomics for cancer targeting and metabolic engineering –A review. Process Biochemistry. 2020;99:112-122. https://doi.org/10.1016/j.procbio.2020.08.023
- Shi L, Zhang N. Applications of solution NMR in drug discovery. Molecules. 2021;26(3):576. https://doi.org/10.3390/molecules26030576
- Tukulula M, Olasupo IA, Mugumbate GC, et al. Synthesis, stereochemistry and in vitro STD NMR and in silico HIV-1 PR enzyme-binding potential of MBH-derived inhibitors. Journal of Molecular Structure. 2022;1268:133716. https://doi.org/10.1016/j.molstruc.2022.133716