Protein Evolution Services
Creative Biostructure provides Protein Evolution service within a R&D project. Each project will be designed individually to suit the needs of our customers and described in a detailed project report.
Protein evolution is closely tied to the changes and selection of DNA polymorphisms and mutations owing to protein sequence change responding to alteration in DNA sequence. So far, great efforts on understanding protein evolution have aimed at domains, independently folding units from which modern proteins are formed. The most powerful tool for characterizing protein sequences is protein sequence comparison and sequence similarity searching.
Our protein evolution services include:
- Consultation to find the best strategy to evolve the protein (random, rational, or combined approach)
- Cloning & expression using our proprietary cloning technology and production strains
- Sophisticated high throughput screening assay development for desired protein activities and their applications
- Design, generation and screening of the mutant libraries
- Optional: purification and characterization of best performing variants (kinetic properties, selectivity, stability etc.
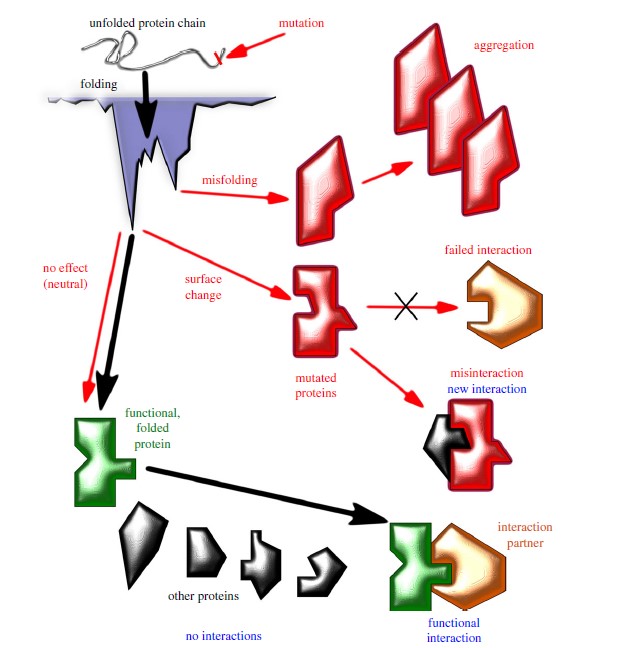
Figure 1. Schematics of protein evolution
Scientists from Creative Biostructure are able to address the gene/function issues by both bioinformatical and experimental methods. Strikingly, we can use computational processes to estimate new domain types or to detect new instances of known domains in poorly characterized proteins. On the other hand, we can use suitable experimental screens to complement the bioinformatical screens for protein functions and validate some of the bioinformatical findings. The rate of protein evolution can be obtained via calculating the evolutionary distance between sequences that are derived from a common ancestor. Evolutionary distances are measured as the number of substitutions per site. In order to estimate distances, it is very necessary to identify orthologous proteins, and to align those amino-acid positions that are derived from the same ancestral site.
Protein evolution requires two steps:
- the mutation of nucleotides that code for amino acids,
- the fixation of new variants in the population.
The probability of fixation depends on the fitness effect of mutations. Three measurements for protein evolution:
- Estimation from amino-acid sequences;
- Estamition from nucleotide suquences;
- Estamition from condon sequences.
Our opitmal strategies for protein evolution services include but not limited to:
- Library design and construction
- Phylogenetic tree construction
- De novo Design
- Directed Evolution
- Rational Design
Creative Biostructure are capable of providing specialized mutant library services, including site-directed mutagenesis libraries, sequential permutation scanning libraries, and randomized and degenerated libraries.
Please feel free to contact us for a detailed quote.
Ordering Process
References:
- J. Söding, and A. N. Lupas (2003). More than the sum of their parts: On the evolution of proteins from peptides. Bioessays 25: 837-846.
- Molecular evolution. (https://en.wikipedia.org/wiki/Molecular_evolution#Protein_Evolution).
- T. Sikosek and H. S. Chan (2014). Biophysics of protein evolution and evolutionary protein biophysics. J. R. Soc. Interface, 11(100): 20140419.
- W. R. Pearson (2001). Protein sequence comparison and Protein evolution. Tutorial - ISMB2000.
