Saturation Transfer Difference (STD) NMR Service
Nuclear magnetic resonance (NMR) is an important screening tool for studying biomolecule-ligand interactions and provides additional structural information compared to commonly used quantitative research methods such as UV-visible spectroscopy, fluorescence spectroscopy, and Fourier transforms infrared spectroscopy.
Among the various NMR techniques, saturation transfer difference (STD) NMR is a screening method that belongs to ligand-based screening. It can identify the binding epitope of the ligand when it binds to the receptor protein and can be used to screen the interaction of small molecules and low molecular weight fragments with a given macromolecule. It is one of the most widely used methods due to its advantages of information diversity, ease of implementation, lower sensitivity to false positives than other screening techniques, reduced protein consumption, and no upper limit on protein size.
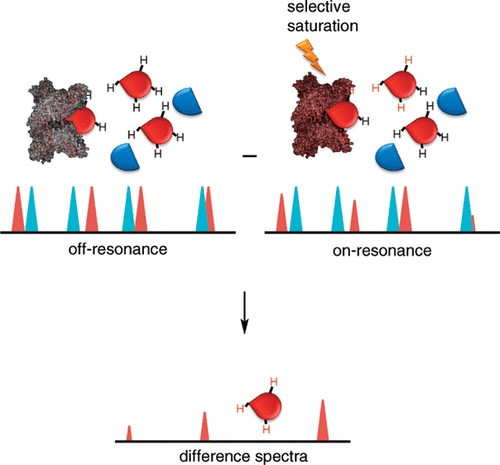 Figure 1. The schematic of saturation transfer difference (STD) NMR. (Viegas, A.; et al. 2011)
Figure 1. The schematic of saturation transfer difference (STD) NMR. (Viegas, A.; et al. 2011)
Creative Biostructure provides technical services based on the STD NMR method to obtain richer information on kinetic and thermodynamic parameters, including mapping of ligand binding epitopes, dissociation constant determination, and binding site properties. This information is crucial in the field of drug discovery and in understanding fundamental biological interactions.
- Detecting ligand-polymer interactions
- Exploring interactions between small molecules (including amino acids) and nanoparticle surfaces
Advantages of Our Technique
- Only a small amount and no isotopically labeled protein is required, and there is no upper limit on the relative molecular mass size of the protein.
- Only signals from the ligand-bound state can be observed, so there is no need to correct for the contribution of the free state. This is particularly advantageous for experimental designs with high ligand-acceptor ratios that might otherwise complicate interpretation.
- By analyzing the relative intensity of the STD NMR signal for each proton in the same compound, ligand epitope mapping can be constructed, providing information on the relative position of the ligand within the binding site.
- Epitope mapping of ligands bound to target proteins at the atomic level can be obtained in the absence of crystalline protein-ligand complexes.
- Easy to implement, and with high sensitivity, can be used to calculate the KD of interactions in the millimolar to micromolar range.
The sensitivity of STD spectra depends on the size of the protein, the duration of the irradiation, the dissociation rate of the ligand, and the degree of excess of the ligand. Therefore, before performing an STD NMR experiment, our scientists design a tailored experimental strategy based on the specific research objectives. Our standard operating procedures ensure the reliability of the experimental data. We will provide all raw data and analysis reports. Please feel free to contact us to start your analysis project as soon as possible.
Ordering Process
Reference
- Viegas A.; et al. Saturation-transfer difference (STD) NMR: a simple and fast method for ligand screening and characterization of protein binding. Journal of Chemical Education. 2011, 88(7): 990-994.
