Analysis of Amyloid Fibrils
Amyloid fibrils are a linear protein aggregate, insoluble in water and resistant to protease activity. Depending on the proteins that make up the fiber, amyloid structures can accumulate in different parts of the body, including the brain, joints, and pancreas. As an expert in the field of nuclear magnetic resonance, Creative Biostructure provides customers with structural, physical, chemical, and biological characteristics analysis services of amyloid fiber based on solid nuclear magnetic resonance.
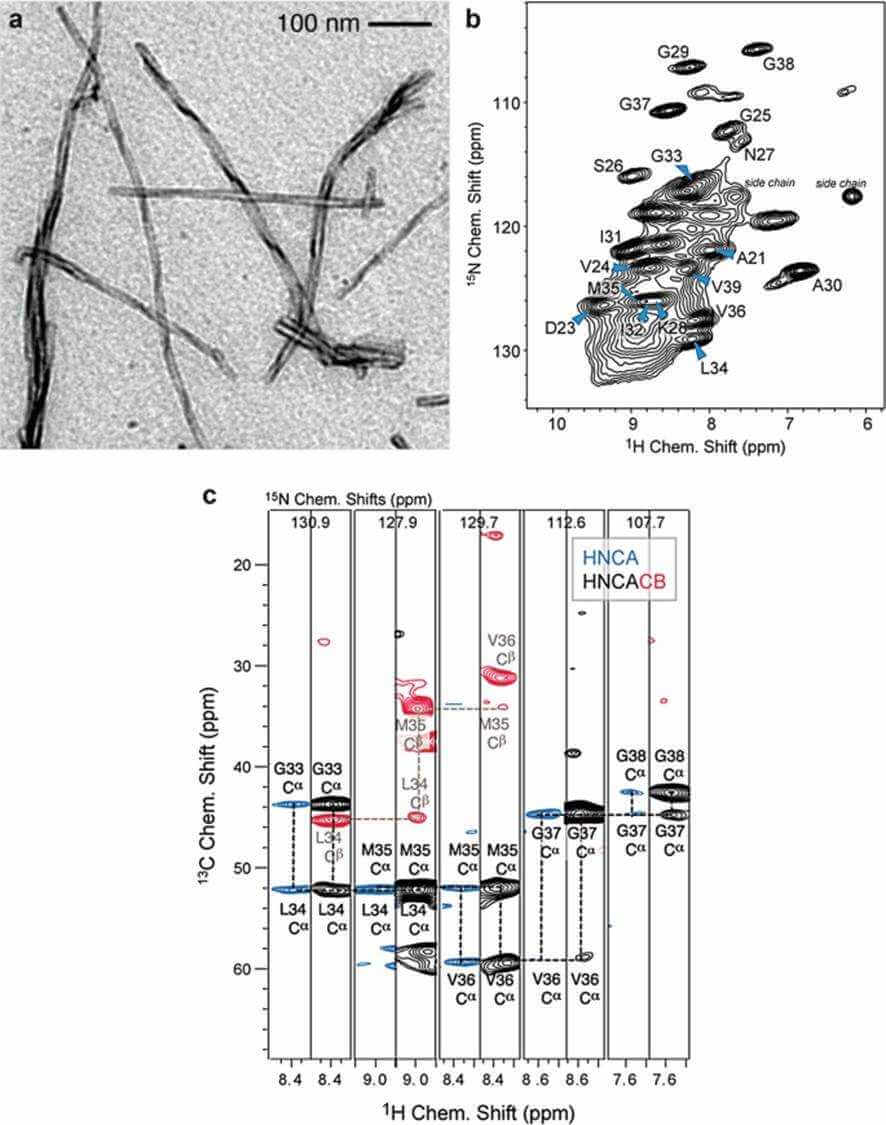 Figure 1. MAS solid-state NMR experiments recorded for a perdeuterated Alzheimer's disease peptide Aβ fibrils (Reif, 2018)
Figure 1. MAS solid-state NMR experiments recorded for a perdeuterated Alzheimer's disease peptide Aβ fibrils (Reif, 2018)
Our Service Purpose
The accumulation of amyloid fibers in tissues is associated with many diseases, including Alzheimer's disease, Parkinson's disease, type 2 diabetes and prion disease. The study of amyloid fibrils can help to understand the etiology, prevention, and treatment of amyloid diseases.
Amyloid fibrils are usually stable structural states of polypeptides. Therefore, another goal of amyloid fibrils research is to clarify the physical and chemical factors that stabilize the structure of amyloid protein and affect the assembly mechanism of fibrils.
Analysis Content of Amyloid Fibrils
- Our structural analysis service for amyloid fiber mainly includes three aspects.
- Conformation of a single protein molecule.
- Fiber structure formed by protein molecules and molecular assembly.
- The intermediate state in the process of protein molecules forming fibers.
Our Approaches
Multi dimension correlation experiment for signal attribution
NMR analysis of protein structure first requires signal attribution, that is, the signal in the NMR spectrum corresponds to a specific atom in the protein molecule to obtain the chemical shift information of each atom. Our commonly used solid-state NMR multidimensional correlation experiments are as follows.
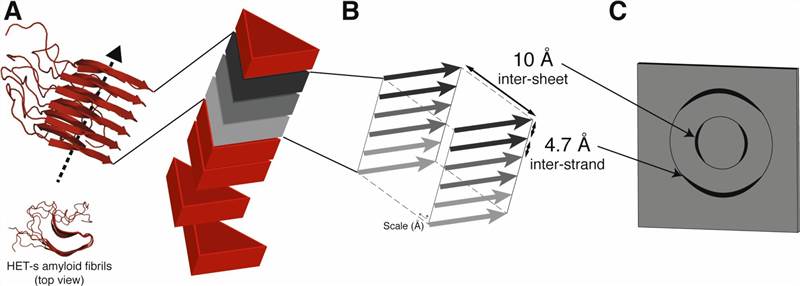 Figure 2. 3D structure determination of amyloid fibrils using solid-state NMR spectroscopy (Loquet et al., 2018)
Figure 2. 3D structure determination of amyloid fibrils using solid-state NMR spectroscopy (Loquet et al., 2018)
| Dimension | Type of experiment |
| 2D | CC,NCA,NCO,N(CA)CX,N(CO)CX |
| 3D | CCC,NCACX,NCOCX,NCACB,CONCA |
| 4D | CANCOCX,CONCACX |
Distance constraint
Analyzing the three-dimensional structure of proteins requires a lot of structural constraints: distance and angle
| Object | Description |
| Homonuclear distance | It is mainly used to measure the distance between isolated spin pairs and plays an important role in selectively labeling proteins. |
| Heteronuclear distance | The distance between heteronuclear 13C-15N was measured by REDOR and TEDOR. In the (13C, 15N) full labeling system, 3D REDOR and 3D TEDOR can provide multiple distance information at the same time. |
Torsion angle constraint
Correlate the interaction of two anisotropy to obtain the torsional angle. Our method includes,
- Correlated CSA dipole coupling
- Dipole coupling - Dipole coupling
- CSA-CSA
Technical Advantages of Solid-State Nuclear Magnetic Resonance
Solid state NMR study of amyloid fiber system has unique advantages, it does not need to crystallize protein and is not limited by aggregate size. It can obtain a three-dimensional structure with atomic resolution, provide global or local dynamic information on fibers, and track the process of fibrosis.
Creative Biostructure is committed to providing high-quality NMR analysis services to advance the life sciences fields. If you have any questions or needs, please contact us and our customer service staff will help you the first time.
Ordering Process
References
- Reif B. Proton-detection in biological MAS solid-state NMR spectroscopy. Modern Magnetic Resonance. 2018: 879-910.
- Loquet A, et al. 3D structure determination of amyloid fibrils using solid-state NMR spectroscopy. Methods. 2018. 138: 26-38.
