Dynamics and Disorder Analysis
Dynamic properties of biomacromolecules are essential for understanding the structural basis of their biological functions such as catalysis, interaction, regulation and cellular signaling. NMR spectroscopy stands for a powerful technique for describing these fundamental features. Supported by our NMR platform, Creative Biostructure is proud to offer services in research of protein dynamics, developing experimental NMR approaches for conformational analysis, and characterizing reorganizational protein disorder.
Protein dynamics affects a wide range of functions, such as catalytic turnover of enzymes, signaling/regulation, binding via induced fit or conformational selection. NMR dynamics is divided into 2 regimes: fast and slow. Fast timescale dynamics ranges from picoseconds to nanoseconds, and is limited by rotational correlation time of protein; slow timescale dynamics spans from microseconds to milliseconds, and requires chemical shift difference. Many interesting biological processes, such as transportation and catalysis, employ slow dynamics. Small domain movements in protein-ligand interaction involve slow-intermediate conformational changes, which can be detected by NMR. (Figure. 1)
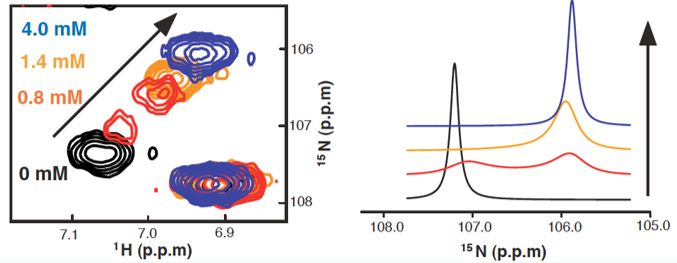 Figure 1. Titration of ligand into protein. Left, Chemical shift undergoing the transition from free to bound state. Right, 1D lineshape simulation of the spectrum.
Figure 1. Titration of ligand into protein. Left, Chemical shift undergoing the transition from free to bound state. Right, 1D lineshape simulation of the spectrum.
NMR spectroscopy offers unique opportunities for structural and dynamic studies of Intrinsically Disordered Proteins (IDPs), which attract a lot of attention recently due to their importance in protein interaction networks in eukaryotic cells. IDPs features flexible natures, which allow them with enormous potential to interact with multiple binding partners, and thus, to efficiently engage in weak regulatory networks. 2D HSQC NMR is exploited for disordered protein structural studies. Disordered protein has much less spreading out spectrum than folded proteins (Fig. 2); therefore, by collecting of a simple NMR spectrum, one can obtain structural information of the target protein.
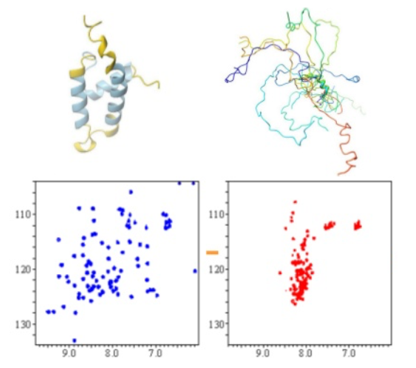 Figure 2. NMR spectra of a protein in its folded and unfolded states
Figure 2. NMR spectra of a protein in its folded and unfolded states
The NMR-based approaches we used for dynamic and disorder studies of protein includes, but not limited to:
- Real-time NMR
- Exchange spectroscopy
- Lineshape analysis
- Rotating frame relaxation dispersion
- Nuclear spin relaxation
- Residual dipolar coupling
- Paramagnetic relaxation enhancement
Please contact us for detailed inquiry!
Ordering Process
References
- Klechner I., Foster M., (2011). An introduction to NMR-based approaches for measuring protein dynamics. Biochim Biophys Acta. 1814 (8): 942-968.
- Wolf-Watz M., Thai V., Henzler-Wildman K., Hadjipavlou G., Eisenmesser E and Kern D., (2004). Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nature structural & molecular biology. 11: 945-949.
