Cryo-EM for Biological Tissues
Conventional electron microscopy techniques have been crucial for studying the structure of biological specimens such as cells, tissues and macromolecules. But sample preparation of conventional electron microscopy requires complete dehydration which leads to material aggregation. Cryo-Electron Microscopy (Cryo-EM) has overcome these limitations and has recently undergone a quantum leap progression in the resolution. Due to it makes the samples without chemical fixations and heavy-metal stains, Cryo-EM has become a powerful technique in structural biology of functional complexes in their native state. Even though without crystallization, and only small amounts of sample are needed, Cryo-EM requires that samples must be very thin in order to acquire interpretable images from electron microscopes.
Cryo-Electron Tomography (Cryo-ET) is one of the techniques of Cryo-EM and it also limited by the thickness of the specimen. Advanced in sample preparation methods and electron microscope technologies, Cryo-ET has become an important method for structural study of tissues, cells, organelles and macromolecular complexes, which makes it possible for people to view cells and tissues not only in their physiological state, but also in their natural environment in the cell.
For years, Creative Biostructure has been devoted to optimizing the Cryo-EM protocol and combining with the advanced methods of high-pressure frozen and freeze-substituted. At Creative Biostructure, We have a professional team for making use of advanced tools of Cryo-ultramicrotomy and Cryo-FIB-SEM (Cryo Focused-Ion-Beam Scanning Electron Microscopy) to thin samples. We are dedicated to provide its customers with high-resolution imaging and three-dimensional (3D) reconstruction services for the biological tissues.
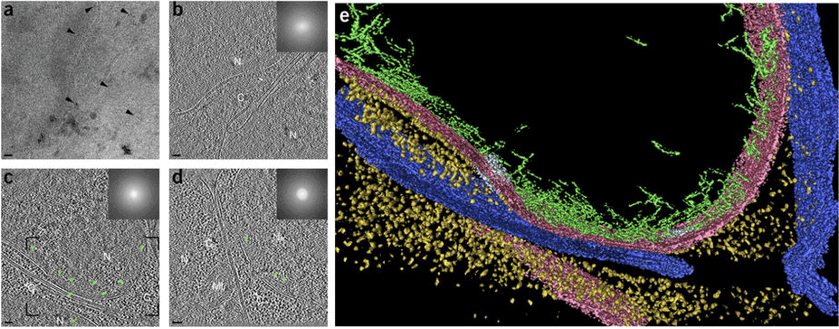 Figure 1. Cryo-ET of vitrified C. elegans embryos.
Figure 1. Cryo-ET of vitrified C. elegans embryos.
Creative Biostructure provides professional services of Cryo-EM. It is possible to observe the structure of biological tissues in the native state and help to define essential biological functions.
Please feel free to contact us for a detailed quote.
Ordering Process
References
- Jan Arnold, Julia Mahamid, et al. Site-Specific Cryo-focused Ion Beam Sample Preparation Guided by 3D Correlative Microscopy. Biophys J. 2016;110:860-869.
- Jan Harapin, Mandy Börme, et al. structural analysis of multicellular organisms with cryo-electron tomography. Nat Methods, 2015;12:634-636.
- Chyongere Hsieh, Thomas Schmelzer, et al. Practical workflow for cryo focused-ion-beam milling of tissues and cells for cryo-TEM tomography. J Struct Biol. 2014; 185:32-41.
- Mark S. Ladinsky, Micromanipulator-Assisted Vitreous Cryosectioning and Sample Preparation by High-Pressure Freezing. Methods Enzymol, 2010; 481:165-194.

