Structural Research of Phosphotransferases
Phosphate transferases (PTs) catalyze phosphorylation reactions, with receptors typically consisting of alcohol, carboxyl, nitrogen-containing, and phosphate groups. The most typical PTs are kinases that catalyze the transfer of a phosphate group from a high-energy donor molecule, such as ATP, to a specific target molecule. PTs are essential components of an organism's energy metabolism. Therefore, exploring the structure of PT is significant for further exploration of its involvement in various cellular processes, including cellular respiration, glycolysis, and the citric acid cycle.
Methods and significance of structural studies on PTs
Taking B. cereus aminoglycoside phosphotransferase (APH) as an example, a 3D model of APH is constructed, and the stability of the APH model is confirmed through molecular dynamics (MD) simulation. Use DSSP diagrams to demonstrate the untwisted behavior of secondary structural elements (spirals and sheets in all APH models). The APH model can further explore the molecular-level mechanisms of drug resistance and serve as a target for drug discovery. Therefore, elucidating the structure of APH is significant in combating bacterial antibiotic resistance at an early stage.
Research on the structure of LcpA
LcpA is a phosphotransferase that mediates glycosylation of Gram-positive bacterial cell wall anchoring proteins. As shown by X-ray crystallography, LcpA folds in an α-β-α structure containing conserved catalytic arginine residues and stable disulfide bonds. Seven strands of antiparallel β-sheets form the core of the protein, and a total of eight α-helices flank the β-sheets to form hydrophobic tunnels. The structure provides more evidence that LcpA possesses pyrophosphatase and PT activities.
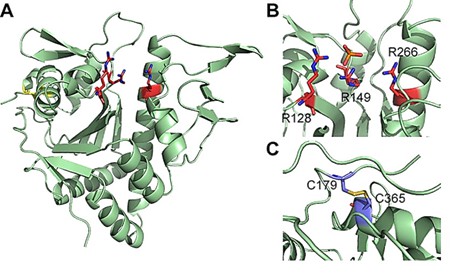 Figure 1. Crystal structure of LcpA (A) and detailed view of the active site (B) and disulfide bonds (C). (Siegel, S. D., et al., 2019)
Figure 1. Crystal structure of LcpA (A) and detailed view of the active site (B) and disulfide bonds (C). (Siegel, S. D., et al., 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Phosphoinositide 3-kinase | Sus scrofa | X-ray diffraction | 2.2 Å | 1E8X |
| Phosphoinositide 3-kinase | Sus scrofa | X-ray diffraction | 2 Å | 1E7U |
| GLKwith aminopyrrolopyrimidine inhibitor | Homo sapiens | X-ray diffraction | 2.85 Å | 5J5T |
| The SH3 domain of MLK3 in complex with poly-proline peptide derived | Homo sapiens | X-ray diffraction | 2 Å | 6CQ7 |
| Unbound SH3 domain of MLK3 | Homo sapiens | X-ray diffraction | 1.5 Å | 5K28 |
| Deoxyribonucleoside Kinase | Drosophila melanogaster | X-ray diffraction | 2.56 Å | 1J90 |
| Deoxyribonucleoside kinase activates gemcitabine in transduced cancer cell lines | Drosophila melanogaster | X-ray diffraction | 2.2 Å | 2VPP |
| Deoxyribonucleoside kinase in complex with DTTP | Drosophila melanogaster | X-ray diffraction | 2.4 Å | 1OE0 |
| DLK (kinase domain) | Homo sapiens | X-ray diffraction | 1.7 Å | 5CEN |
| Pyrimidine nucleoside phosphorylase in a closed conformation | Geobacillus stearothermophilus | X-ray diffraction | 2.1 Å | 1BRW |
| Thymidine phosphorylase | Escherichia coli K-12 | X-ray diffraction | 2.6 Å | 2TPT |
| Thymidine phosphorylase | Escherichia coli K-12 | X-ray diffraction | 1.55 Å | 4EAF |
| Thymidine phosphorylase complex with SO4 | Salmonella enterica subsp. enterica serovar Typhimurium | X-ray diffraction | 2.2 Å | 4X46 |
| Polyphosphate kinase | Escherichia coli | X-ray diffraction | 3 Å | 1XDO |
| Polyphosphate Kinase in complex with AMPPNP | Escherichia coli | X-ray diffraction | 2.5 Å | 1XDP |
| Thiamin pyrophosphokinase | Bacteroides thetaiotaomicron VPI-5482 | X-ray diffraction | 1.8 Å | 2OMK |
| HPPK(H115A) | Escherichia coli K-12 | X-ray diffraction | 1.996 Å | 3KUG |
| The small alarmone synthetase 2 | Bacillus subtilis subsp. subtilis str. 168 | X-ray diffraction | 3.2 Å | 6FGK |
| Phosphorybosyl pyrophosphate synthetase II | Thermus thermophilus | X-ray diffraction | 1.85 Å | 7PN0 |
Table 1. Structural research of phosphotransferases.
Creative Biostructure epitomizes excellence and expertise in the field of structural research. Our expert team possesses unrivaled access to cutting-edge technologies such as NMR spectroscopy, cryo-electron microscopy (cryo-EM) and X-ray crystallography, which allow us to gain a deep and comprehensive understanding of the intricate structure and function of membrane proteins.
We pride ourselves on delivering high-quality, accurate, and timely structural analysis results that set us apart from our competitors. If you would like to delve deeper into the structure of phosphotransferases and other proteins, then please do not hesitate to contact us. No matter how challenging your research needs may be, our expert team is always ready to discuss and explore with you and provide you with the best solutions and strategies for your project.
References
- Siegel, S. D., et al. Structure and Mechanism of LcpA, a Phosphotransferase That Mediates Glycosylation of a Gram-Positive Bacterial Cell Wall-Anchored Protein. mBio, 2019.10(1): e01580-18.
- Parulekar, R.S., Sonawane, K.D. Structure elucidation study of aminoglycoside phosphotransferase from B. cereus sensu lato: a comprehensive outlook for drug discovery. Struct Chem. 2023.34:859–865.
- Boyko KM, et al. Structural characterization of the novel aminoglycoside phosphotransferase AphVIII from Streptomyces rimosus with enzymatic activity modulated by phosphorylation. Biochem Biophys Res Commun. 2016.477(4):595-601.
