Structural Research of Peptidases
Peptidases, also known as proteolytic enzymes, can hydrolyze peptide chains and be divided into two main types, namely endopeptidases, and exopeptidases. Endopeptidase acts on specific peptide bonds within the peptide chain. In contrast, exopeptidase acts on the N-terminal or C-terminal of the peptide substrate, catalyzing the generation of free amino acids and small peptides.
The signal peptide is a membrane-bound proteolytic enzyme, which plays a crucial role in bacterial survival by processing transmembrane translocation proteins. Signal peptide peptidase A (SppA) is a membrane-bound self-compartmentalized serine protease in plants, archaea, and bacteria.
Scientists reported the crystal structure of the catalytic domain of Bacillus subtilis SppA (SppABS) with a resolution of 2.40 Å. The analysis results of X-ray crystallography revealed an octamer complex composed of eight SppABS molecules. The active site of SppABS is formed through the assembly of octamer, and the adjacent protoplasts each provide half of the catalysis. It also describes the substrate-specific pocket of SppABS and points out the difference between SppABS and Escherichia coli SppA (SppAEC) in the range of peptides they can lyse.
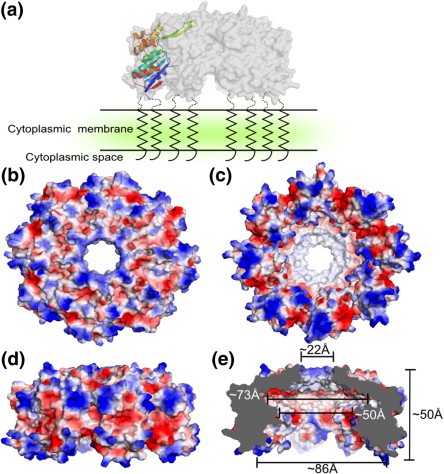 Figure 1. Surface properties and dimensions of SppABS. (NAM, et al., 2012)
Figure 1. Surface properties and dimensions of SppABS. (NAM, et al., 2012)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| LepB Signal Peptidase (SPase) in complex with a β-lactam inhibitor | Escherichia coli BL21(DE3) | X-ray diffraction | 1.95 Å | 1B12 |
| SPase | Escherichia coli K-12 | X-ray diffraction | 2.40 Å | 1KN9 |
| LepB Signal Peptidase (SPase) in complex with a lipopeptide inhibitor | Escherichia coli, Streptomyces sp. | X-ray diffraction | 2.47 Å | 1T7D |
| SpsB Signal Peptidase (SPase) | Escherichia coli K-12, Staphylococcus aureus subsp. aureus str. Newman | X-ray diffraction | 2.05 Å | 4WVG |
| SPase in complex with peptide 1 | Escherichia coli K-12, Staphylococcus aureus subsp. aureus str. Newman, Staphylococcus aureus | X-ray diffraction | 2.10 Å | 4WVH |
| SPase in complex with peptide 2 | Escherichia coli K-12, Staphylococcus aureus subsp. aureus str. Newman, Staphylococcus aureus | X-ray diffraction | 1.90 Å | 4WVI |
| SPase in complex with peptide 3 | Escherichia coli K-12, Staphylococcus aureus subsp. aureus str. Newman, Staphylococcus aureus | X-ray diffraction | 1.95 Å | 4WVJ |
| Signal Peptide Peptidase (SppA), native protein | Escherichia coli | X-ray diffraction | 2.55 Å | 3BF0 |
| SeMet protein | Escherichia coli | X-ray diffraction | 2.76 Å | 3BEZ |
| Signal Peptide Peptidase (SppA) | Bacillus subtilis | X-ray diffraction | 2.37 Å | 3RST |
| Signal Peptide Peptidase (SppA) K199A mutant showing C-terminal peptide bound in eight active sites | Bacillus subtilis subsp. subtilis str. 168 | X-ray diffraction | 2.39 Å | 4KWB |
| Meprin β sheddase | Homo sapiens | X-ray diffraction | 1.85 Å | 4GWM |
| Meprin β sheddase, mature form | Homo sapiens | X-ray diffraction | 3.00 Å | 4GWN |
Table 1. Structural research of peptidases.
X-ray crystallography has important application value in analyzing complex biomolecules such as membrane proteins. Creative Biostructure's X-ray crystallography service is committed to providing technical support for research in life science. Our professional team will work with you to customize the most suitable solution for your research needs. We use high-intensity X-ray and precise detectors to obtain high-resolution structural data, ensuring that our customers can obtain the most reliable and accurate results. We are committed to providing you with the best service and helping you achieve further breakthroughs in biomolecular research.
References
- AROLAS J L, et al. Structural basis for the sheddase function of human meprin β metalloproteinase at the plasma membrane. Proceedings of the National Academy of Sciences, 2012, 109(40): 16131–16136.
- NAM S-E, et al. Structure of signal peptide peptidase A with C-termini bound in the active sites: Insights into specificity, self-processing, and regulation. Biochemistry, 2013, 52(49): 8811–8822.
- NAM S-E, et al. Crystal structure of bacillus subtilis signal peptide peptidase a. Journal of Molecular Biology, 2012, 419(5): 347–358.
- KIM A C, et al. Crystal structure of a bacterial signal peptide peptidase. Journal of Molecular Biology, 2008, 376(2): 352–366.
- TING Y T, et al. Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization. IUCrJ, 2016, 3(1): 10–19.
- PAETZEL M, et al. Crystallographic and biophysical analysis of a bacterial signal peptidase in complex with a lipopeptide-based inhibitor. Journal of Biological Chemistry, 2004, 279(29): 30781–30790.
- PAETZEL M, et al. Crystal structure of a bacterial signal peptidase apoenzyme. Journal of Biological Chemistry, 2002, 277(11): 9512–9519.
- PAETZEL M, et al. Crystal structure of a bacterial signal peptidase in complex with a β-lactam inhibitor. Nature, 1998, 396(6707): 186–190.