Structural Research of Cytochromes P450
Cytochrome P450 enzyme is a widely present heme containing monooxygenase, involved in the synthesis of steroid hormones, lipid soluble vitamin metabolism, polyunsaturated lipids preventing acid conversion into bioactive molecules, as well as carcinogenic effects and drug metabolism.
Cytochrome P450 is an oxidoreductase that catalyzes the transfer of oxygen atoms using thiol salt bound heme as the active center. It requires heme iron (HEM) to activate oxygen The iron ion of the active center HEM can chelate with cysteine sulfur on one side and can chelate with oxygen molecules in water on the other side The absolutely conserved cysteine in cytochrome P450 forms a thiol ion bond with the iron element in the catalytic active center heme, becoming a ligand for iron.
The crystal structure of dozens of cytochromes P450 shows significant sequence differences among different species, but its three-dimensional structure is highly conserved All cytochrome P450 are based on sequences that maintain similarity in the folding and bending of the entire structure, and their secondary structures are labeled using typical cytochrome P450 folding nomenclature.
Combining the structures of several cytochrome P450, it was found that the heme group was limited between the terminal helix I and the adjacent helix L. Cysteine near the catalytic center forms two hydrogen bonds with adjacent amino groups in the main chain. The long helix I forms the inner wall of the heme pocket and contains the characteristic amino acid sequence. Threonine, which is highly conserved and has acidic residues, is located at the active site and participates in catalysis.
Lanosterol 14-α-demethylase or CYP51 enzyme, probably the oldest enzyme in cytochrome P450 family, plays a central role in cholesterol or ergosterol biosynthesis. This enzyme can catalyze the first post cyclization step in ergosterol biosynthesis and is inhibited by triazole drugs. These structures reveal an ordered N-terminal amphiphilic helix located before the assumed transmembrane helix, which will limit the direction of the catalytic domain and partially locate in the lipid bilayer.
The X-ray crystal structure of full-length Saccharomyces cerevisiae 14-α-demethylase (ScErg11p-6xHis) has been clarified. Its membrane domain is structurally distinguishable, and there are two ligand crystals: the abstract lanosterol and the triazole inhibitor ITC. The clear electron density of 50 N-terminal amino acids shows two helices with mutual orientation of~60°. The N-terminal helix [membrane helix 1 (MH1), residues 6-23] is amphiphilic and has extensive crystal contact with symmetry related molecules. The distribution of hydrophobic and hydrophilic side chains indicates that the hydrophobic surface may contact the inner lobule of endoplasmic reticulum (ER) membrane. MH1 is connected to TMH1 through a short helix containing proline (residues 24-26), which is a 37.5 Å long slightly twisted helix (residues 27-51) long enough to pass through the lipid bilayer. The side chains of amino acids R30 and Q29 adjacent to the turn form the C-terminal cap of helix MH1 and the N-terminal cap of TMH1, respectively.
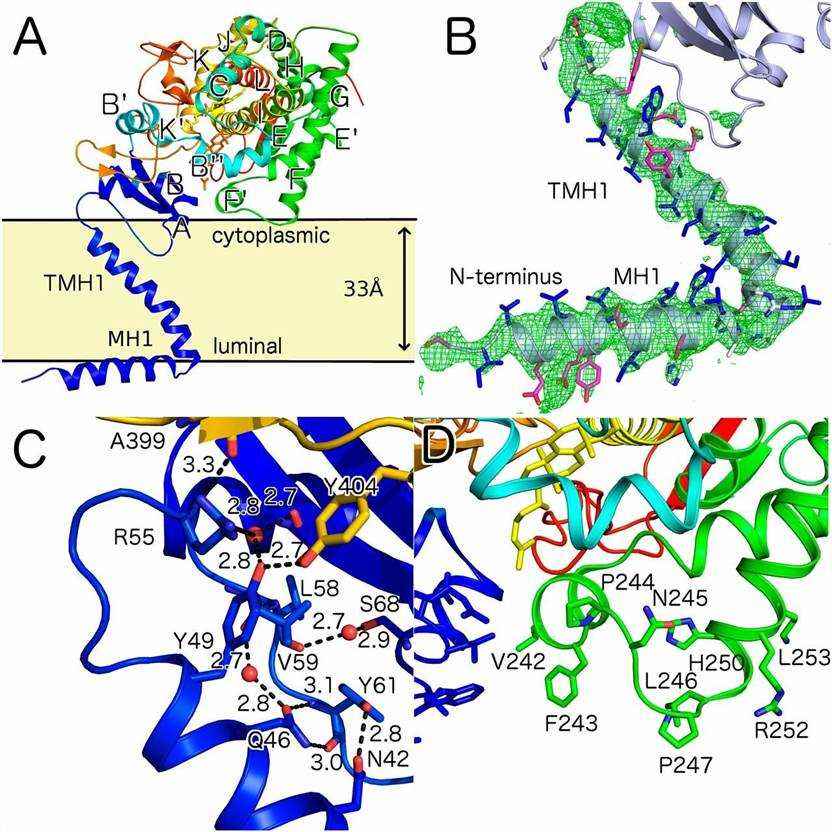 Figure 1. Overall fold of ScErg11p and predicted orientation in the lipid bilayer. (Monk B C, et al., 2014)
Figure 1. Overall fold of ScErg11p and predicted orientation in the lipid bilayer. (Monk B C, et al., 2014)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Lanosterol 14α-demethylase with bound lanosterol | Saccharomyces cerevisiae | X-ray diffraction | 1.90 Å | 4LXJ |
| Lanosterol 14α-demethylase with bound itraconazole | Saccharomyces cerevisiae | X-ray diffraction | 2.19 Å | 5EQB |
Table 1. Structural research of Cytochromes P450.
As a leader in structural biology, Creative Biostructure has high-quality equipment and extensive experimental experience in X-ray crystallography, and our services provide powerful tools for studying the three-dimensional structure of membrane proteins. If you are interested in learning more about our protein structural analysis services, please contact us for more information.
Reference
- Monk B C, et al. Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proceedings of the National Academy of Sciences, 2014, 111(10): 3865–3870.