Structural Research of Phosphoenolpyruvate-Dependent Phosphotransferases (PTSs)
The phosphoenolpyruvate-(PEP)-dependent phosphotransferase system (PTS) is the main carbohydrate uptake system in bacteria. This protein phosphorylation chain can recognize extracellular signals and intracellular signals (such as phosphoenolpyruvate and nitrogen), transferring phosphoenolpyruvate to transported carbohydrates. Studying the structure of the proteins in this system allows further investigation of the enzymatic activity and action mechanism in catalytic reactions.
Composition and functional studies of phosphoenolpyruvate-(PEP)-dependent PTS
Almost all phosphoenolpyruvate-(PEP)-dependent PTSs contain two cytoplasmic phosphotransferases, enzyme I (EI) and histidine phosphate carrier protein (HPr), as well as glycan-specific enzyme II complexes (EIIA, EIIB, EIIC, EIID). PTS transports and phosphorylates carbohydrates. It is also involved in carbon metabolism regulatory functions such as catabolic inhibition and inducer deterrence. In addition to controlling nitrogen metabolism, regulating iron or potassium homeostasis, and the virulence of certain pathogens, it mediates stress responses to nutrient stress and physicochemical stimuli.
Research progress in bcMalT structure
The bcMalT is a member of the glucose superfamily of EIIC membrane-bound enzymes. The bcMalT's crystal structure is captured in an inward-facing conformation by an Hg2+ that bridges two cysteine residues. The structure permits direct comparison of outward- and inward-facing conformations and reveals large rigid-body motions of the sugar-binding structural domains and other conformational changes that accompany rigid-body motions. Molecular dynamics (MD) simulations are also used to test that the bcMalT cross-linked (bcMalT-X) structure is stable.
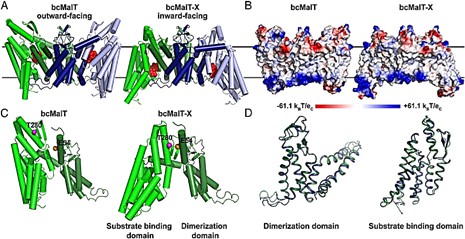 Figure 1. Structural comparison of bcMalT and bcMalT-X. (Ren Z, et al., 2018)
Figure 1. Structural comparison of bcMalT and bcMalT-X. (Ren Z, et al., 2018)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Phosphoenolpyruvate-dependent phosphotransferase system | Bacillus subtilis | X-ray diffraction | 2.9 Å | 1BLE |
| Inward-facing conformation of L-ascorbate transporter UlaA | Pasteurella multocida | X-ray diffraction | 3.333 Å | 5ZOV |
| bcMalT T280C-E54C crosslinked by divalent mercury | Bacillus cereus E33L | X-ray diffraction | 3.2 Å | 6BVG |
| The full-length Enzyme I of the PTS system | Staphylococcus carnosus | X-ray diffraction | 2.5 Å | 2HRO |
| man-PTS | Listeria monocytogenes | Cryo-EM single particle analysis | 3.12 Å | 7VLX |
| man-PTS complexed with pediocin PA-1 | Listeria monocytogenes | Cryo-EM single particle analysis | 2.45 Å | 7VLY |
| The transporter ChbC | Bacillus cereus ATCC 10987 | X-ray diffraction | 3.295 Å | 3QNQ |
| The transporter MalT | Bacillus cereus E33L | X-ray diffraction | 2.551 Å | 5IWS |
| Vitamin C transporter UlaA/SgaT in P21 form | Escherichia coli K-12 | X-ray diffraction | 2.359 Å | 4RP8 |
| Vitamin C transporter UlaA/SgaT in C2 form | Escherichia coli K-12 | X-ray diffraction | 1.651 Å | 4RP9 |
| Mannose transporter ManYZ and microcin E492 (MceA) complex | Escherichia coli K-12 Klebsiella pneumoniae |
Cryo-EM single particle analysis | 2.28 Å | 7DYR |
| Gluconate-specific EIIA phosphotransferase system component | Enterococcus faecalis | X-ray diffraction | 2.5 Å | 3IPR |
| Glucose permease IIA | Mycoplasma capricolum | X-ray diffraction | 2.5 Å | 2GPR |
| IIA mannitol | Escherichia coli K-12 | X-ray diffraction | 1.8 Å | 1A3A |
| IIB cellobiose | Escherichia coli K-12 | X-ray diffraction | 1.8 Å | 1IIB |
| IIA domain of the glucose permease | Bacillus subtilis | X-ray diffraction | 1.9 Å | 1GPR |
| 6-phospho-beta-glucosidase | Lactiplantibacillus plantarum | X-ray diffraction | 1.498 Å | 3QOM |
| 6-phospho-beta-glucosidase mutant (E375Q) in complex with Salicin 6-phosphate | Streptococcus mutans | X-ray diffraction | 2.54 Å | 4F79 |
| 6-phospho-beta-glucosidase in complex with beta-D-glucose-6-phosphate. | Streptococcus mutans | X-ray diffraction | 1.479 Å | 4F66 |
| Solution structure of the N-terminal membrane anchor of enzyme IIA(Glucose) | Escherichia coli | SOLUTION NMR | / | 1O53 |
| Enzyme GatB of the galactitol-specific phosphoenolpyruvate-dependent phosphotransferase system | Escherichia coli | SOLUTION NMR | / | 1TVM |
| The mannitol-specific cryptic phosphotransferase enzyme IIA CmtB | Escherichia coli | SOLUTION NMR | / | 2OQ3 |
Table 1. Structural research of phosphoenolpyruvate-dependent phosphotransferases (PTSs).
The significance of structural analysis services in the study of phosphoenolpyruvate-dependent phosphotransferases (PTSs) cannot be underestimated. At Creative Biostructure, we offer various structural analysis services to help researchers better understand protein structure and function. We offer a variety of techniques such as NMR spectroscopy, cryo-electron microscopy (cryo-EM) and X-ray crystallography that allow us to accurately determine protein structure.
We develop services tailored to our client's specific research needs. Our team of senior scientists has extensive experience working with phosphoenolpyruvate-dependent phosphotransferases (PTSs) and other complex proteins, and we have an unwavering commitment to providing unparalleled structural analysis services. If you are looking to realize your research goals, contact us to learn more about our structural analysis services.
References
- Ren Z, et al. Structure of an EIIC sugar transporter trapped in an inward-facing conformation. Proc Natl Acad Sci U S A. 2018.115(23):5962-5967.
- Morabbi Heravi K, Altenbuchner J. Cross talk among transporters of the phosphoenolpyruvate-dependent phosphotransferase system in bacillus subtilis. J Bacteriol. 2018.200(19): e00213-18.
- Cao Y, et al. Crystal structure of a phosphorylation-coupled saccharide transporter. Nature. 2011.473(7345):50-54.