Structural Research of Papillomaviridae
Papillomaviridae is a group of small, enveloped, dsDNA viruses that are classified based on paired nucleotide sequence identity within the L1 open reading frame. These viruses primarily infect mucosal and keratinized epithelial cells and can lead to the development of lesions. Human papillomavirus (HPV) is the most typical member of this family, and infection with it usually causes benign epithelial lesions (warts), and some types of HPV have been implicated as the causative agent of cervical and anal cancers. In recent years, researchers have used cryo-electron microscopy (cryo-EM) to reconstruct the structure of Papillomaviridae members, essential for understanding their assembly, entry, pathogenesis, and tumorigenesis. This has paved the way for the development of targeted interventions and improved public health outcomes associated with papillomavirus infection and related diseases.
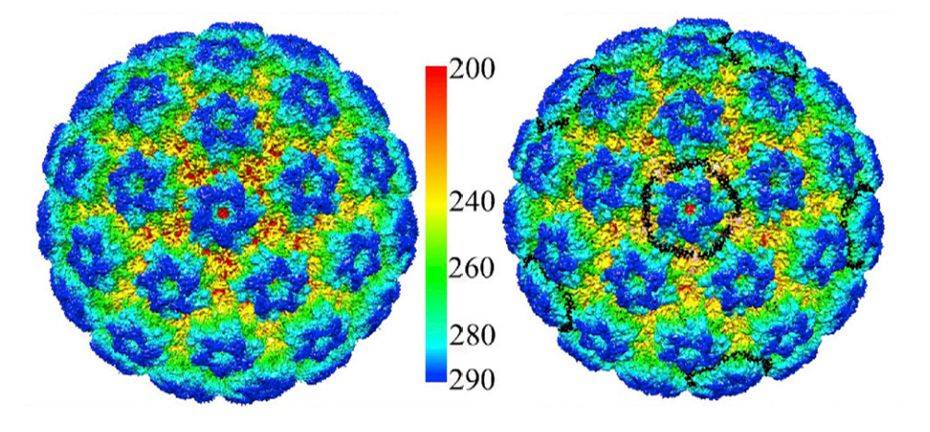 Figure 1. Cryo-EM Reconstruction of HPV16 and HPV16-Heparin. (Guan J, et al, 2017)
Figure 1. Cryo-EM Reconstruction of HPV16 and HPV16-Heparin. (Guan J, et al, 2017)
Papillomavirus Genome Structure
All HPVs contain a double-stranded circular DNA genome of about 8kb in size, which contains about eight open reading frames (ORFs), all of which are transcribed from a single DNA strand. The ORFs can be divided into an early (E) region (encoding the protein E1-E7 required for virus replication); a late-stage (L) region (encoding the structural protein L1-L2 required for virus particle assembly); and an upstream regulatory region (including cis elements necessary for viral DNA replication and transcription). Of these, the E1 and E2 proteins play a major role in replication, while the L1 and L2 proteins are involved in the assembly of the viral genome.
Structural Information on HPV Viral Particles
HPV is an envelope-less, circular dsDNA-containing virus with particles of approximately 55nm in diameter. Its capsid consists of 360 copies of the major capsid protein L1 and the minor structural protein L2. Five copies of L1 are intertwined to form 72 capsids constituting a T = 7 d icosahedron, which exhibits significant differences between HPV types. Recently, the HPV16 VLP capsids were researched by cryo-electron microscopy (cryo-EM) and image analysis. The structures showed that the VLP is stabilized by a disulfide bond between L1, with a ring protruding to the outer surface containing epitopes recognized by the immune system. The L1 exhibits three different structural states, the L1 monomer, the L1 pentamer, and an icosahedral structure consisting of 12 pentamers. Of these, the icosahedral VLP is the most effective in triggering an immune response.
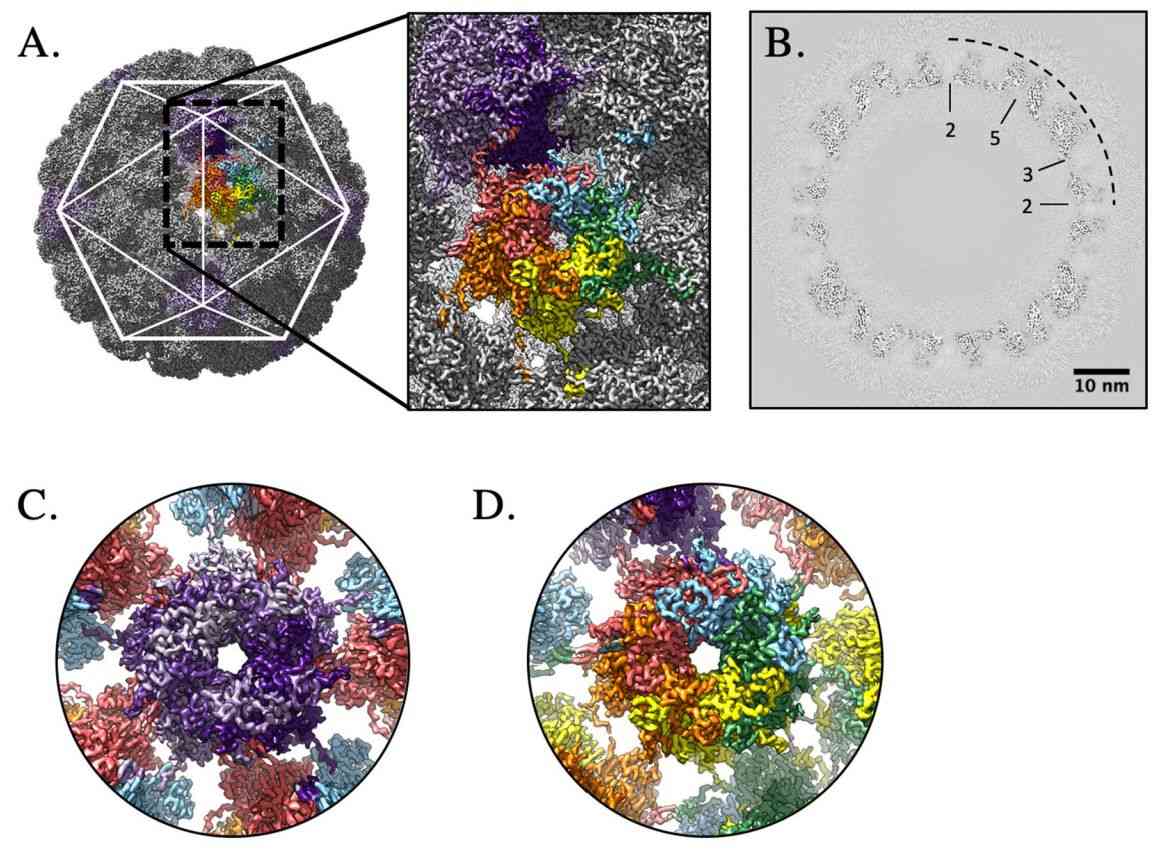 Figure 2. High-resolution capsid map of HPV16. (Goetschius DJ, et al, 2021)
Figure 2. High-resolution capsid map of HPV16. (Goetschius DJ, et al, 2021)
Cervical cancer through HPV infection is the second most common cancer in women worldwide. To date, there are no appropriate drug molecules for the treatment of HPV infection, and the identification of novel anti-HPV potential drugs with unique modes of action is essential.
Creative Biostructure recognizes that structure elucidation is essential in the discovery of HPV vaccine and antiviral drugs. We provide high-purity virus-like particles (VLPs) for viral structural analysis to help clients understand the mechanism of viral entry into host cells and the interactions between viral proteins and the host for the development of targeted therapies.
| Cat No. | Product Name | Virus Name | Source | Composition |
| CBS-V074 | HPV VLP | Human papillomavirus | Yeast recombinant | Surface antigen |
| CBS-V553 | Bovine papillomavirus 1 VLP (L1 Proteins) | Bovine papillomavirus | E. coli recombinant | L1 |
| CBS-V637 | HPV16 VLP (L1 Proteins) | Human papillomavirus | Yeast recombinant | L1 |
| CBS-V639 | Chimeric HPV16 VLP (L1-E7(1–57) Proteins) | Human papillomavirus | Insect cell recombinant | L1-E7(1–57) |
| Explore All Papillomaviridae Virus-like Particle Products | ||||
Creative Biostructure is dedicated to the structural analysis of biological macromolecules. We provide professional structure analysis services for viruses and virus-like particles based on cutting-edge cryo-electron microscopy (cryo-EM) technologies. We provide the best customer service using advanced methods to help clients drive breakthrough discoveries in virology.
Our extensively trained and certified laboratory staff have the expertise and proficiency to ensure accurate and reliable data delivery. Contact us to experience our professional, efficient, and quality service.
References
- Guan J, et al. Cryoelectron Microscopy Maps of Human Papillomavirus 16 Reveal L2 Densities and Heparin Binding Site. Structure. 2017. 25(2): 253-263.
- Goetschius DJ, et al. High resolution cryo EM analysis of HPV16 identifies minor structural protein L2 and describes capsid flexibility. Sci Rep. 2021. 11(1): 3498.
- Van Doorslaer K, et al. ICTV Virus Taxonomy Profile: Papillomaviridae. J Gen Virol. 2018. 99(8): 989-990.