Structural Research of Nucleobase-Cation-Symport-2 (NCS2) Family
The nucleobase-cation-symport-2 (NCS2) family is also known as the nuclear base ascorbate transporter (NAT) family. It comes from gram-negative and Gram-positive bacteria, archaea, fungi, plants, and animals. In bacteria and fungi, the NCS2 protein mainly transports uracil, xanthine, and other purines or pyrimidine derivatives. In mammals, SVCT1 and SVCT2 in the family are localized to different tissues of the organism and co-transport L-ascorbic acid and Na+.
Research Progress of NCS2/NAT Protein Structure
UraA is a representative NAT protein. The UraA is a folded structure with 14 transmembrane segments (TMs) divided into two inverted repeats. A pair of antiparallel β chains, located between TM3 and TM10, play an important role in structural organization and substrate recognition. Uracil is located at the interface between the two domains, and the transport function is mainly performed by residues of the core domain.
NCS2/NAT Protein Structure Research Applications
Purines and pyrimidine analogues are widely used against a variety of diseases and infections. The study of the relationship between the structure and function of NAT can be used to design purine-related antifungals. These antifungals are not metabolized by the host's transport system. Kinetic studies are carried out and the binding model of the drug and substrate is proposed. Such a model can be used in the design of nucleobase-related analogs. NATs do not use N3 to bind uric acid and xanthine. N3-substituted purine analogues are directed into nucleic acid synthesis and can act as antiviral inhibitors at low concentrations.
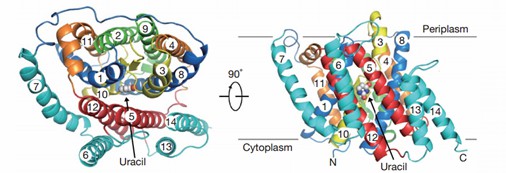 Figure 1. UraA view from the ectoplasm and the side. (Lu, F., et al., 2011)
Figure 1. UraA view from the ectoplasm and the side. (Lu, F., et al., 2011)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Uracil Transporter--UraA | Escherichia coli K-12 | X-ray diffraction | 2.781 Å | 3QE7 |
| The eukaryotic purine/H+ symporter, UapA, in complex with Xanthine | Aspergillus nidulans FGSC A4 | X-ray diffraction | 3.7 Å | 5I6C |
| UraA in occluded conformation | Escherichia coli O157:H7 | X-ray diffraction | 2.5 Å | 5XLS |
| The xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | Bacillus subtilis | X-ray diffraction | 1.32 Å | 4FE5 |
| The AU25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | Bacillus subtilis | X-ray diffraction | 1.6 Å | 4FEO |
| The A24U/U25A/A46G/C74U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with 2,6-diaminopurine | Bacillus subtilis | X-ray diffraction | 1.65 Å | 4FEP |
| The A24U mutant xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | Bacillus subtilis | X-ray diffraction | 1.5 Å | 4FEG |
| The A24U/U25A/A46G mutant xpt-pbuX guanine riboswitch aptamer domain in complex with hypoxanthine | Bacillus subtilis | X-ray diffraction | 3.00 Å | 4FEN |
| Xpt-pbuX C74U Riboswitch bound to acetoguanamine identified by virtual screening | Bacillus subtilis | X-ray diffraction | 1.59 Å | 2XNZ |
| Xpt-pbuX C74U Riboswitch bound to N6-methyladenine | Bacillus subtilis | X-ray diffraction | 1.6 Å | 2XO1 |
| Guanine riboswitch A21U, U75A mutant bound to hypoxanthine | Guanine riboswitch | X-ray diffraction | 1.75 Å | 2EES |
Table 1. Structural research of nucleobase-cation-symport-2 (NCS2) family.
To study the structure of nucleobase-cation-symport-2 (NCS2) family proteins, X-ray crystallography is commonly used. Structural analysis of membrane proteins facilitates further research of their functions and mechanisms.
Creative Biostructure has long been committed to the study of structural biology and membrane proteins. Our structural analysis services are not limited to membrane proteins, we can accurately analyze biomolecules, including but not limited to nucleic acids, ribosomes, small proteins, protein complexes, protein-ligand complexes, and viruses. If you are interested in our services, please contact us and we will provide you with a professional and comprehensive solution.
References
- Lu, F., et al. Structure and mechanism of the uracil transporter UraA. Nature, 2011;472: 243–246.
- Gournas C, et al. The nucleobase-ascorbate transporter (NAT) family: genomics, evolution, structure-function relationships and physiological role. Mol Biosyst. 2008;4(5):404-416.
- Funk, A.M., et al. A nucleobase cation symporter 2, EaXanP, from Erwinia amylovora transports xanthine. J Plant Pathol, 2021;103,89–98.