Structural Research of Monotopic Decarboxylases
Decarboxylases are a group of enzymes that remove the carboxyl group (CO2H) from acidic substrates and require pyridoxal phosphate or pyruvate as cofactors. Decarboxylases are well known for their various roles in metabolic pathways and carbohydrate synthesis.
PS decarboxylase (PSD) can synthesize phosphatidylethanolamine (PE), one of the most abundant membrane phospholipids in prokaryotes and eukaryotes, through the decarboxylation reaction of phosphatidylserine (PS).
The study of the PSD structure helps to elucidate the catalytic and substrate recognition mechanisms of this enzyme. The crystal structures of the apolipoprotein form and the PE-bound form of PSD (EcPsd) from E. coli can be determined by X-ray crystallography at 2.6 and 3.6 Å resolution, respectively. EcPsd forms a homodimer with each protomer having a positively charged substrate binding pocket at the active site. Structure-based mutational analysis indicates that conserved residues in the pocket are involved in PS decarboxylation. ecPsd has an N-terminal hydrophobic helix region that is important for membrane binding, resulting in efficient PS recognition.
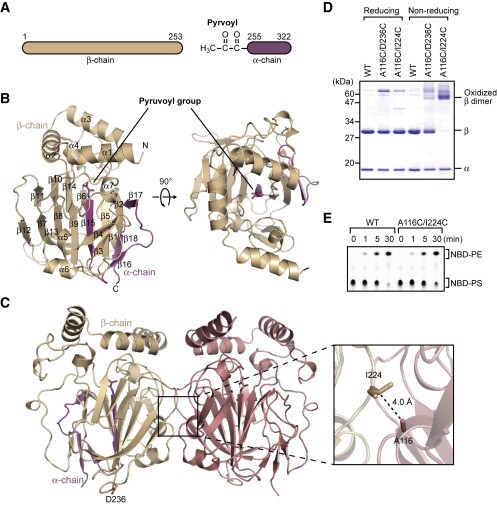 Figure 1. Structure of EcPsd. (Watanabe Y, et al., 2020)
Figure 1. Structure of EcPsd. (Watanabe Y, et al., 2020)
The asymmetric unit of the EcPsd crystal contains four EcPsd molecules with similar conformations to each other and a root-mean-square deviation (RMSD) of 0.4 Å. The α and β chains of each EcPsd monomer in the asymmetric unit form a spherical fold consisting of seven α helices and 18 β chains (Figure 1B). A β sandwich structure with nine β chains on one side and four β chains on the other side forms in the center of the fold and is surrounded by four short α helices (α4-α7) and five β chains. The three N-terminal helices (α1-α3) are located above the β sandwich structure, forming a "U" shape. In the asymmetric unit, the four EcPsd molecules form two homodimers (Figure 1C), each monomer adopting a new fold with a positively charged substrate binding pocket at the center of the fold. The acetonyl cofactor covalently bound to the N-terminal end of the EcPsd α chain is located at the bottom of the pocket. the headgroup portion of the PE molecule is accommodated in the pocket and forms a reduced Schiff base bond with the acetonyl cofactor. Structure-based mutational analysis indicates that the conserved Tyr and His residues in the pocket play an important role in PS decarboxylation. ecPsd binds to the membrane through the N-terminal hydrophobic helix region around the pocket, resulting in efficient PS recognition.
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Phosphatidylserine decarboxylase (PSD), apo-form | Escherichia coli BL21(DE3) | X-ray diffraction | 2.60 Å | 6L06 |
| Phosphatidylserine decarboxylase (PSD), PE-bond | Escherichia coli BL21(DE3) | X-ray diffraction | 3.60 Å | 6L07 |
| Phosphatidylserine decarboxylase (PSD), apo-form | Escherichia coli K-12 | X-ray diffraction | 1.90 Å | 7CNW |
| Phosphatidylserine decarboxylase (PSD), apo-form | Escherichia coli K-12 | X-ray diffraction | 2.63 Å | 7CNX |
| Phosphatidylserine decarboxylase (PSD), with 8PE bound | Escherichia coli K-12 | X-ray diffraction | 2.12 Å | 7CNY |
| Phosphatidylserine decarboxylase (PSD), with 10PE bound | Escherichia coli K-12 | X-ray diffraction | 2.70 Å | 7CNZ |
| Oxaloacetate Decarboxylase Sodium Pump (OAD) βγ sub-complex | Salmonella enterica subsp. enterica serovar Typhimurium | Cryo-EM structure | 3.90 Å | 6IWW |
| Oxaloacetate Decarboxylase Sodium Pump (OAD) βγ sub-complex | Salmonella enterica subsp. enterica | X-ray diffraction | 4.40 Å | 6IVA |
Table 1. Structural Research of Decarboxylases.
Creative Biostructure offers best-in-class X-ray crystallography and cryo-electron microscopy (cryo-EM) services, powerful tools for studying the three-dimensional structure of membrane proteins. We use the latest assay equipment and techniques to provide you with the best possible service and results. Our specially trained and qualified laboratory personnel have the experience and skills to provide you with the most reliable and accurate data. If you are interested in learning more about our protein structural analysis services, please contact us for more information.
References
- Watanabe Y, et al. Structural basis for phosphatidylethanolamine biosynthesis by bacterial phosphatidylserine decarboxylase. Structure, 2020, 28(7).
- Cho G, et al. Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis. Scientific Reports, 2021, 11(1).
- XU X, et al. Structural insights into sodium transport by the oxaloacetate decarboxylase sodium pump. eLife, 2020, 9.