Structural Research of Sec, Translocase, and Insertase Proteins
Protein translocation and membrane integration are fundamentally conserved processes. After or during protein synthesis, precursor proteins containing N-terminal signaling sequences are directed to a conserved membrane protein complex called Sec transposon. Sec transposon consists of the Sec61 complex in eukaryotic cells or the SecY complex in bacteria. Therefore, structural analysis of the Sec transposon can deepen the understanding of the structure-function relationship of translocation and its interaction with auxiliary components during protein translocation and membrane protein insertion.
Structural elucidation of the channel complex SecYEG
The heterotrimeric protein complex SecYEG is a central player in protein translocation. Reconstruction studies with purified SecYEG complexes have shown that the minimal translocase consists of the SecYE complex and the motor protein SecA. The crystal structure of SecYEβ from Methanocaldococcus jannaschii provides the first high-resolution insights into the organization and structure of the translocation channel. The SecY protein consists of 10 α-helical TMSs, with pseudo-symmetric arrangements of TMSs 1-5 and TMSs 6-10. SecE wraps around the SecY channel in a V-shape and contacts the two SecYs by tilted helices and amphipathic helices, respectively. "shells". These two helices are connected by a hinge region that provides flexibility to the structure. The Secβ subunit is located further out in the structure.
Progress in structural research on the ribosome- Sec transposon complex
In recent years, researchers have investigated the structure of ribosome- Sec transposon complexes by medium-resolution electron microscopy, revealing the density of α-helices and conformational changes in the transmembrane region, which provide new insights into protein transmembrane translocation and integration. Electron microscopy images of the ribosome-nascent chain complex (RNC) include various intermediate states of the SecY complex, thus providing snapshots of several co-translational translocations. cryo-electron microscopy structures of Sec61 and the RNC show that the signaling peptides are viewed similarly to the secye - seca signaling peptide complex. Analysis of different cryo-EM imaging of SecY and RNC showed that two transmembrane regions are located near the outer gate of SecY.
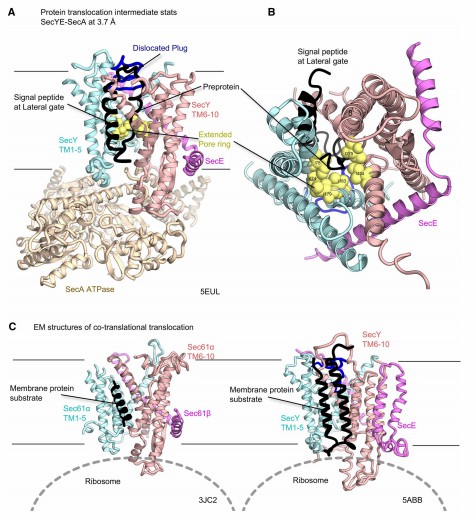 Figure 1. Structures of the Sec translocon in the intermediate stages of protein transport. (Tsukazaki T, 2019)
Figure 1. Structures of the Sec translocon in the intermediate stages of protein transport. (Tsukazaki T, 2019)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Bacterial holo-translocon | Escherichia coli | Cryo-EM single particle analysis | 14 Å | 5MG3 |
| YidC | Escherichia coli | X-ray diffraction | 2.8 Å | 6AL2 |
| YidC | Thermotoga maritima MSB8 |
X-ray diffraction | 3.842 Å | 5Y83 |
| Twin Arginine Translocase Receptor- TatC | Aquifex aeolicus VF5 | X-ray diffraction | 4 Å | 4HTS |
| TatAd protein in DPC micelles | Bacillus subtilis | SOLUTION NMR | / | 2L16 |
| The periplasmic domain of YidC | Thermotoga maritima MSB8 | X-ray diffraction | 2.517 Å | 5Y82 |
| TatA T22P | Escherichia coli K-12 | SOLUTION NMR | / | 2LZR |
| TatA oligomer | Escherichia coli K-12 | SOLUTION NMR | / | 2LZS |
| YidC (form I) | Halalkalibacterium halodurans C-125 | X-ray diffraction | 2.403 Å | 3WO6 |
| YidC (form II) | Halalkalibacterium halodurans C-125 | X-ray diffraction | 3.201 Å | 3WO7 |
| Active YidC with the first transmembrane domain resolved | Thermotoga maritima MSB8 | X-ray diffraction | 3.4 Å | 6Y86 |
| SecYE translocon | Thermus thermophilus | X-ray diffraction | 6 Å | 2ZQP |
| Peptide-Bound State of SecYEG | Thermus thermophilus HB8 | X-ray diffraction | 3.64 Å | 5CH4 |
| Protein conducting channel | Methanocaldococcus jannaschii | X-ray diffraction | 3.5 Å | 1RHZ |
| The precise resting state of SecYEG | Thermus thermophilus HB8 | X-ray diffraction | 2.724 Å | 5AWW |
| SecYE translocon with a Fab fragment | Mus musculus | X-ray diffraction | 3.2 Å | 2ZJS |
| Ribosome-bound SecYEG translocon | Escherichia coli | Cryo-EM single particle analysis | 6 Å | 6R7L |
| SecDF in I form | Deinococcus radiodurans R1 = ATCC 13939 = DSM 20539 | X-ray diffraction | 4 Å | 5XAM |
| SecDF in Super-membrane-facing form | Thermus thermophilus HB8 | X-ray diffraction | 2.8 Å | 5YHF |
| SecA-SecY complex with a translocating polypeptide substrate | Bacillus subtilis subsp. subtilis str. 168 | X-ray diffraction | 3.7 Å | 5EUL |
| SecDF, a translocon-associated membrane protein | Thermus thermophilus HB8 | X-ray diffraction | 3.3 Å | 3AQP |
Table 1. Structural research of Sec, translocase, and insertase proteins.
Creative Biostructure is proud to offer a full suite of cutting-edge protein structure analysis services. Our X-ray crystallography, cryo-electron microscopy (cryo-EM), and nuclear magnetic resonance (NMR) service can determine the structure of proteins at high resolution and provide protein structures in different conformations.
Our team of scientists has an unparalleled wealth of experience and an unrelenting commitment to excellence and is dedicated to delivering outstanding results in the structural elucidation of Sec, translocase, and insertase proteins. We utilize cutting-edge equipment and techniques to ensure that our results are both accurate and highly reliable. If you are interested in our services, please feel free to contact us. We are ready to provide you with valuable insights into the complex structure and function of Sec transposons.
References
- Tsukazaki T. Structural Basis of the Sec Translocon and YidC Revealed Through X-ray Crystallography. Protein J. 2019. 38(3): 249-261.
- Dong L, et al. Structural basis of SecA-mediated protein translocation. Proc Natl Acad Sci USA. 2023. 120(2): e2208070120.
- Lycklama A Nijeholt JA, Driessen AJ. The bacterial Sec-translocase: structure and mechanism. Philos Trans R Soc Lond B Biol Sci. 2012. 367(1592): 1016-1028.