Structural Research of Membrane-Bound Metalloproteases
Membrane-bound metalloproteases have various functions such as protein hydrolysis, degradation and remodeling of extracellular matrix, and intracellular signal transduction. Most of them contain zinc-bound metalloprotease domains. By regulating signal transduction and tumor microenvironment, it participates in pathological processes including tumor occurrence, development, invasion, and metastasis. Therefore, deeper research into the function of membrane-bound metalloproteases in cancer immune regulation will help develop cancer diagnosis and immunotherapy methods.
Structure and action mechanism of Escherichia coli zinc metalloprotease RseP
RseP from Escherichia coli is a zinc metalloprotease site 2 protease (S2P) homolog. It participates in the second step of signal transduction by sequentially cleaving type II membrane proteins. The Ec RseP structure contains three C-terminal residues. TM1 contains two zinc coordination His residues. TM3 is divided into two segments by a circular protrusion. Ec RseP is expected to have two inner membranes β Hairpin (the C1N loop and MREβ-loop), located between TM1 and TM2.
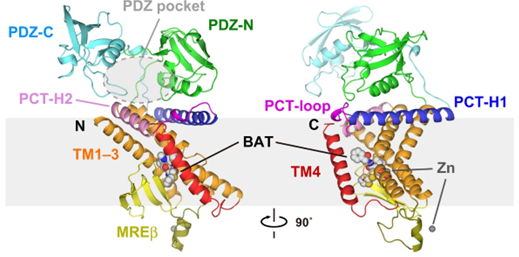 Figure 1. Two views of the full-length Ec RseP structure. (Yuki Imaizumi, et al., 2022)
Figure 1. Two views of the full-length Ec RseP structure. (Yuki Imaizumi, et al., 2022)
Structure Mycobacterium tuberculosis M13 metalloprotease Zmp1
Zmp1 is a type II transmembrane protein with a short N-terminal intracellular structural domain, a transmembrane helix, and a protease structural domain localized in extracellular or intracellular vesicles. It has D1 and D2 structural domains, both consisting of helices. Two open-state cryo-EM structures of Zmp1 have been reported at 3.1 Å and 4.6 Å resolution, and the dominant state in solution is revealed by SAXS analysis.
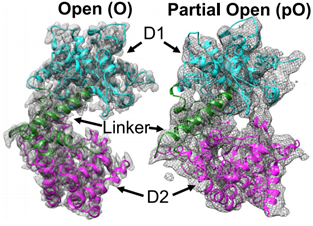 Figure 2. Cryo-EM maps of Zmp1. (Liang WG, et al., 2021)
Figure 2. Cryo-EM maps of Zmp1. (Liang WG, et al., 2021)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| PRO form of proMMP-7 in complex with zwitterionic membrane | Homo sapiens | SOLUTION NMR | / | 2MZH |
| PRO form of proMMP-7 in complex with anionic membrane | Homo sapiens | SOLUTION NMR | / | 2MZI |
| M1 zinc metallopeptidase E323A mutant | Deinococcus radiodurans R1 | X-ray diffraction | 1.83 Å | 6IFF |
| M1 zinc metallopeptidase E323A mutant bound to Tyr-ser-ala substrate | Deinococcus radiodurans R1 | X-ray diffraction | 1.9 Å | 6IFG |
| Two domain M1 Zinc metallopeptidase E323A mutant bound to L-tryptophan amino acid | Deinococcus radiodurans R1 | X-ray diffraction | 2.35 Å | 6KOY |
| Partial open state of zinc metalloprotease 1 | Mycobacterium tuberculosis H37Rv | X-ray diffraction | 4.6 Å | 7K1V |
| RseP orthologue in complex with batimastat in space group P21 | Kangiella koreensis DSM 16069 | X-ray diffraction | 3.15 Å | 7W6Z |
| PDZ-C domain fragment of RseP orthologue | Kangiella koreensis DSM 16069 | X-ray diffraction | 1.15 Å | 7W70 |
| Zinc metalloprotease zmp1 in open state | Mycobacterium tuberculosis H37Rv | X-ray diffraction | 3.1 Å | 6XLY |
| Insulin-regulated aminopeptidase with alanine in active site | Homo sapiens | X-ray diffraction | 3.02 Å | 4P8Q |
| Insulin-regulated aminopeptidase with lysine in active site | Homo sapiens | X-ray diffraction | 2.96 Å | 4PJ6 |
| Insulin-regulated aminopeptidase in complex with ligand | Homo sapiens | X-ray diffraction | 3.31 Å | 4Z7I |
| Insulin regulated aminopeptidase | Homo sapiens | X-ray diffraction | 3.37 Å | 5C97 |
| Insulin-regulated aminopeptidase complexed with a macrocyclic peptidic inhibitor | Homo sapiens | X-ray diffraction | 3.2 Å | 6YDX |
| Aminopeptidase N | Escherichia coli | X-ray diffraction | 1.5 Å | 2DQ6 |
| Aminopeptidase N complexed with bestatin | Escherichia coli | X-ray diffraction | 1.6 Å | 2DQM |
| Aminopeptidase N | Neisseria meningitidis MC58 | X-ray diffraction | 2.05 Å | 2GTQ |
| PepN (Aminopeptidase N)in complex with Bestatin | Escherichia coli K-12 | X-ray diffraction | 2.3 Å | 2HPT |
| Aminopeptidase N in complex with arginine | Escherichia coli K-12 | X-ray diffraction | 2 Å | 3B2P |
| Aminopeptidase N in complex with Lysine | Escherichia coli | X-ray diffraction | 1.5 Å | 3B2X |
Table 1. Structural research of membrane-bound metalloproteases.
To study the structure of membrane proteins, we use NMR spectroscopy, cryo-electron microscopy (cryo-EM) and X-ray crystallography. Structural analysis of membrane-bound metalloproteases contributes to the development of novel antimicrobial targets and cancer therapies.
Creative Biostructure has long been committed to the study of structural biology and membrane proteins. Our experts have extensive experience in determining the structure of membrane proteins. If you are interested in our services, please contact us and we will provide you with a professional and comprehensive solution.
References
- Yuki Imaizumi, et al. Mechanistic insights into intramembrane proteolysis by E. coli site-2 protease homolog RseP. Sci. Adv. 2022. 8, eabp9011.
- Liang WG, et al. Structural analysis of Mycobacterium tuberculosis M13 metalloprotease Zmp1 open states. Structure. 2021. 29(7):709-720.