Antigen-Antibody Complex Structure
In immunology, the antigen (Ag) refers to substances that can induce an immune response in the body, that is, antigens can specifically recognize and bind to the antigen receptor (TCR/BCR) on the surface of T/B lymphocytes, resulting in activation of T/B cells. The activated T/B cells proliferate and differentiate, producing immune response products (sensitized lymphocytes or antibodies), which are capable of specifically binding to the corresponding antigens in vitro and in vivo. Therefore, antigenic substances possess two important characteristics: immunogenicity and immunoreactivity.
Anything that can stimulate the immune system is an antigen. The antigen molecule can be proteins, polysaccharides, nucleic acids, or lipids. Sometimes, the foreign body itself stimulates the immune system. Therefore, the antigen can also be pollen, pathogens, and spores. In autoimmune diseases, the antigen is part of the host itself.
An antibody (Ab) is a large glycoprotein molecule produced primarily by plasma cells and utilized by the immune system to neutralize antigens. It is the main effector molecule of humoral immune response and is found in blood and tissue fluids. They specifically bind or recognize invading pathogenic microorganisms; therefore, they are an essential part of the immune system. The term "antibody" refers to its function, which binds to an antigen. Another name for this protein molecule is immunoglobulin (abbreviated as Ig).
- Antibody Structure
- Antibody Classification
- The Structural Basis of Antigen-Antibody Complex
- Structure Determination of Antigen-Antibody Complex
Antibody Structure
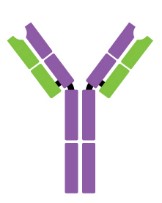
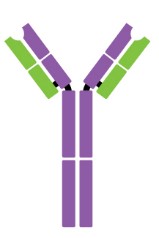
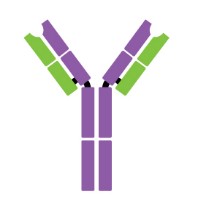
Antibodies are Y-shaped proteins consisting of two identical heavy chains (H chains) and two identical light chains (L chains) joined together by disulfide bonds. The most commonly discussed antibody molecule is immunoglobulin G (IgG). A typical IgG antibody molecule has 12 domains (termed VH, CH1, CH2, CH3, VL, and CL) arranged in two H chains and two L chains linked together by disulfide bonds. Typically, IgG can be cleaved by papain into three separate fragments at the hinge region, two of which are identical and can bind to antigens, known as a fragment of antigen-binding (Fab). The third segment is termed a fragment of crystalline (Fc) because it is more susceptible to crystallization.
The N-terminal domain of each polypeptide (H and L chains) is highly variable, and this portion is referred to as the variable region (V region), which is associated with antigen recognition and determines the specificity of recognition. The remaining domains have constant sequences called the constant region (C region). Additionally, a comparison of V region sequences indicates that the variability is not uniformly distributed but concentrated in areas called hypervariable regions. Because the hypervariable regions are sites where the antibodies direct contact with antigenic epitopes, they are also referred to as complementarity-determining regions (CDRs). The non-hypervariable regions in the V regions have a relatively constant sequence of amino acid residues, and these residues constituting a stable three-dimensional framework structure (known as framework regions) are required to maintain the indispensable immunoglobulin fold leading to the three-dimensional proximity of CDRs. Although the framework regions don't usually form bonds with the antigen, they are necessary for generating the folding of the V region and maintaining the integrity of the binding site.
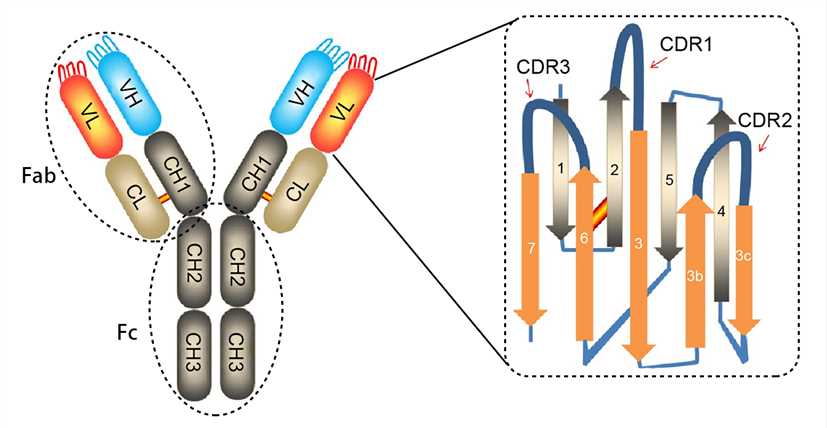 Figure 1. The structure of an antibody molecule.
Figure 1. The structure of an antibody molecule.
Antibody Classification
Based on differences in H chains, mammalian antibodies are classified into five isotypes (IgA, IgD, IgE, IgG, and IgM), which have different distribution and immunological functions. The same Ig isotype can be further classified into subclasses (e.g., IgG is divided into four subclasses IgG1, IgG2, IgG3, and IgG4) by subtle differences in the amino acid composition and the number or position of disulfide bonds. Specific information on each type of antibody is shown in the following table.
| Name | IgA dimer (IgA1, IgA2) | IgD monomer | IgE monomer | IgG monomer (IgG1,2a,2b,3,4) | IgM pentamer (IgM1, IgM2) |
|---|---|---|---|---|---|
| Diagram |
|
|
|
|
|
| Description | With a J chain and a secretory piece. Function in mucosal immunity | Present on the B cell surface as a receptor | Five domains in the H chain; Main participants in allergy | The only type of Ig that have the ability to penetrate the placenta | Early stage immunity |
| Type of H chain | α1, α2 | δ | ε | γ1, γ2, γ3, γ4 | μ |
| Type of L chain | λ, κ | λ, κ | λ, κ | λ, κ | λ, κ |
| Number of antigen-binding sites | 4 | 2 | 2 | 2 | 10 |
| Molecular weight (kDa) | 385 | 180 | 200 | 150 | 900 |
| Percentage of total antibodies in serum | 13% | 1% | 0.002% | 80% | 6% |
Table 1. Types and characteristics of antibodies.
The Structural Basis of Antigen-Antibody Complex
- Antigenic epitope and paratope
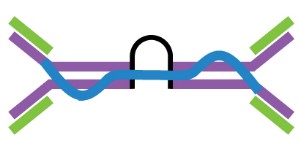
Antigen-antibody interactions are driven by specific contacts between the V regions (H and L chains) and the antigen surface. The specific contact of antigens is referred to as antigenic epitope, and the contact on antibodies is termed paratope.
The antigenic epitopes are generally divided into two main types: linear epitope and conformational epitope. Linear epitopes are formed by a continuous sequence of amino acid residues in a protein, while conformational epitopes are composed of several discrete amino acid sequences, which are widely separated in the primary structure, but are brought together on the surface when the polypeptide chains fold to form the three-dimensionally folded protein. Many therapeutic monoclonal antibodies recognize conformational epitopes of target antigens, which are often represented by membrane proteins, receptors, or multi-subunit proteins.
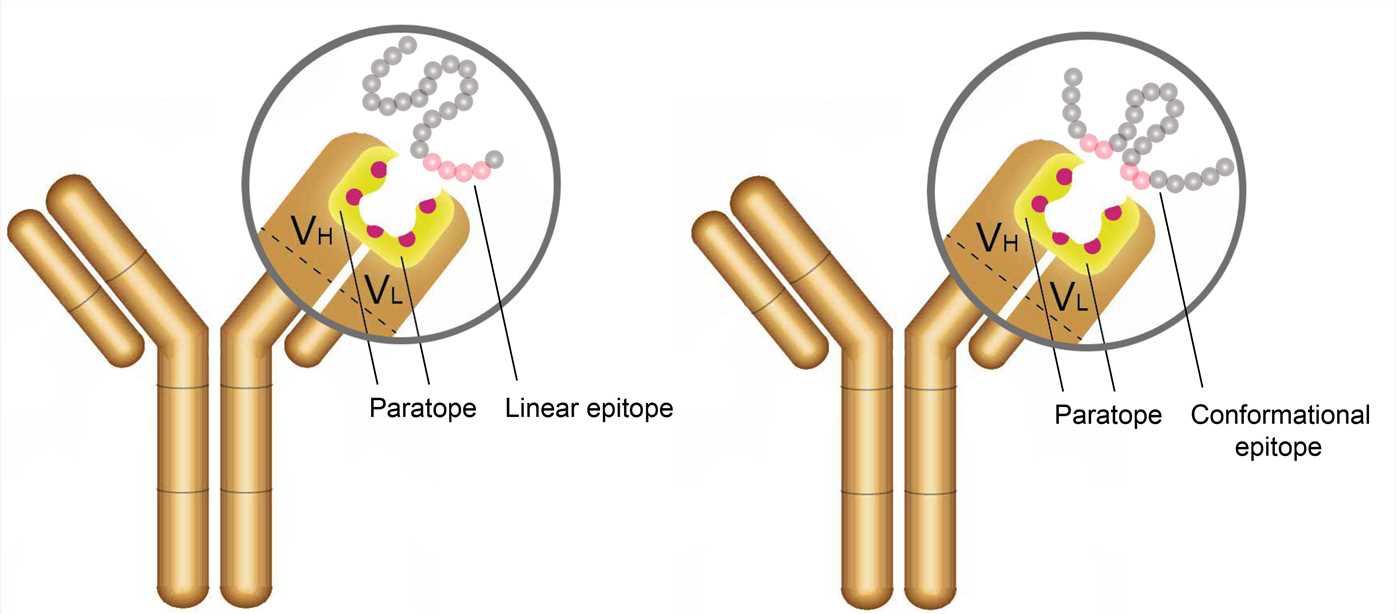 Figure 2. The antigenic epitope and paratope.
Figure 2. The antigenic epitope and paratope.
- Forces involved in antigen-binding
The antigenic epitope interacts with the paratope by the formation of multiple noncovalent bonds, and they must be in close contact with each other in space to generate sufficient binding force. The complementary structure between the molecules determines the specificity of antigen-antibody binding. The strength of a single antigen-antibody bond is affinity. It is produced by the summation of attractive and repulsive forces, which include hydrogen bonds, ionic bonds, hydrophobic interactions, and van der Waals interactions. Minor changes in antigenic structure can significantly affect the strength of antigen-antibody binding. Loss of a single hydrogen bond at the antigen-antibody interface can reduce the strength of the interaction by 1,000 times. All interactions occur in a balance between the attraction and repulsiveness of the contact surface. The strength of antigen-antibody interactions can also be altered by alteration in amino acid residues at the binding sites.
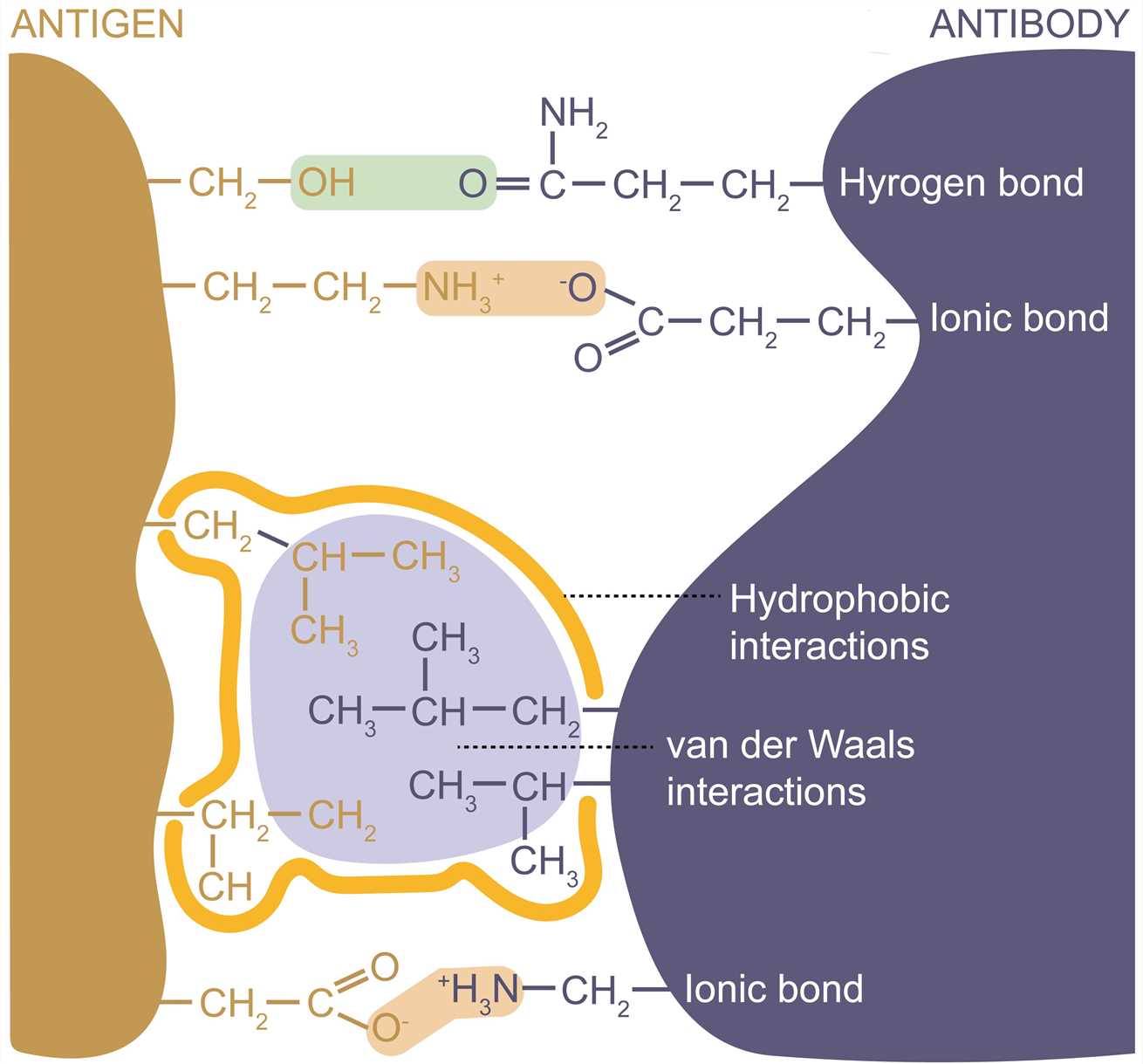 Figure 3. Forces involved in antigen-binding.
Figure 3. Forces involved in antigen-binding.
Structure Determination of Antigen-Antibody Complex
The determined three-dimensional structure of the antigen-antibody complexes allows the identification of the binding sites of antibodies on their target antigens (epitope mapping) and conformational changes induced by binding. Information on the antigen-antibody complex structures is essential for understanding the mechanism of antigen-antibody reaction, as well as for developing the structure-based design of novel therapeutics, vaccines, and diagnostics.
X-ray crystallography provides the most accurate and detailed data on protein structure and interactions. For the past few decades, many three-dimensional structures of antibodies have been successfully obtained by this technology, however, the number of structures determined in complex with their antigens is still relatively small, which is probably because co-crystallization of antigen-antibody complexes remains a major obstacle. Currently, this technology is mainly directed toward the structural determination of Fab-antigen complexes. The reason for choosing a Fab fragment is that it is not as difficult to crystallize as the entire antibody, which has more flexible regions and therefore large conformational heterogeneity. In addition, the determination of the Fab structure is relatively easy.
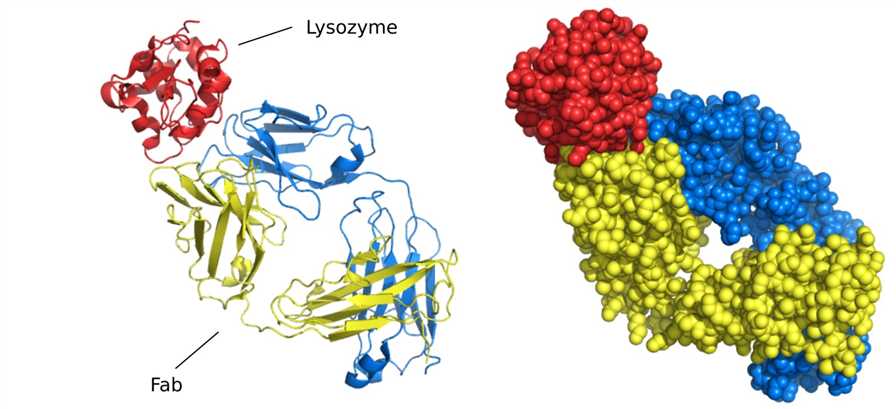 Figure 4. Structure of Fab fragment bound to lysozyme.
Figure 4. Structure of Fab fragment bound to lysozyme.
Cryo-electron microscopy (Cryo-EM) enables the structure determination of large protein complexes. Cryo-EM can circumvent some of the problems of X-ray crystallography and has contributed substantially to the study of antigen-antibody complex structures.
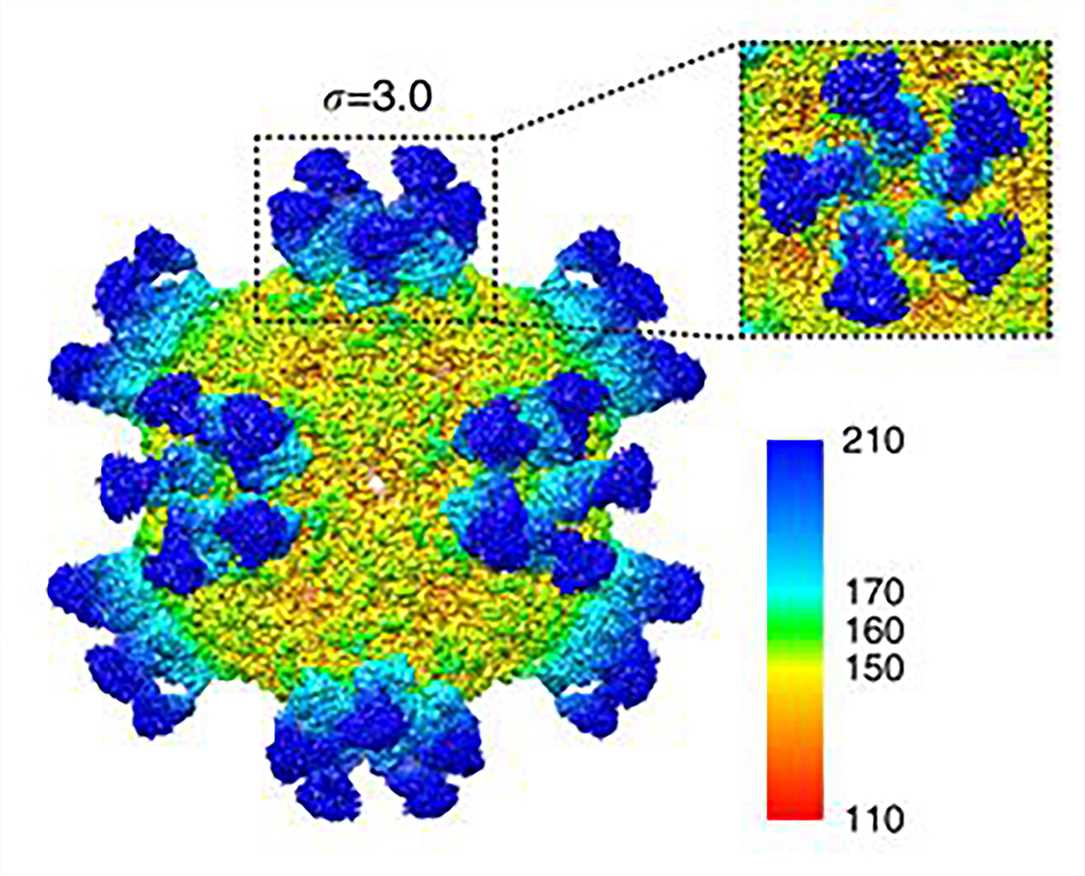 Figure 5. Cryo-EM map of CVA6 A-particle complexed with Fab-1D5.
Figure 5. Cryo-EM map of CVA6 A-particle complexed with Fab-1D5.
Creative Biostructure is specialized in the field of structural biology, and we provide contract services for the structural analysis of antigen-antibody complexes.
References
- Sela-Culang I, et al. The structural basis of antibody-antigen recognition. Frontiers in Immunology. 2013. 4: 302.
- Wang Y, et al. Nanobody-derived nanobiotechnology tool kits for diverse biomedical and biotechnology applications. International Journal of Nanomedicine. 2016. 11: 3287.
- Stura E A, et al. Crystallization of antibodies and antibody-antigen complexes. Immunomethods. 1993. 3(3): 164-179.
- Xu L, et al. Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody. Nature Communications. 2017, 8(1): 1-12.