Structural Research of Major Facilitator Superfamily (MFS) Transporters
The Major Facilitator Superfamily (MFS) is a diverse group of transporters that play crucial roles in cellular physiology by facilitating the translocation of a wide array of substrates across cell membranes.
The Architecture and Transport Mechanism of MFS Transporters
MFS transporters exhibit a conserved architecture consisting of twelve transmembrane helices that form symmetrical N- and C-terminal halves of six gene-duplicated transmembrane units. The central region of the molecule harbors a substrate-binding site, which is pivotal for the translocation of specific molecules across the membrane. The transport mechanism of MFS family members is explained by the "alternating access model." This model postulates that transporter molecules undergo a 'rocker switch' motion between outward-facing, occluded, and inward-facing conformations. This motion facilitates the alternation of a cavity from the binding site to either side of the membrane, enabling substrate transfer across the membrane.
Advancements in Structural Analysis of OxlT
Recent research has furthered our understanding of OxlT's structural basis by utilizing X-ray crystallography. Researchers have successfully determined the X-ray crystallographic structures of OxlT in both oxalate-bound and ligand-free forms, achieving impressive resolutions ranging from 3.0 to 3.3 Å. This higher-resolution structural information provides valuable insights into OxlT's mode of action, specifically its oxalate recognition and antiport mechanism.
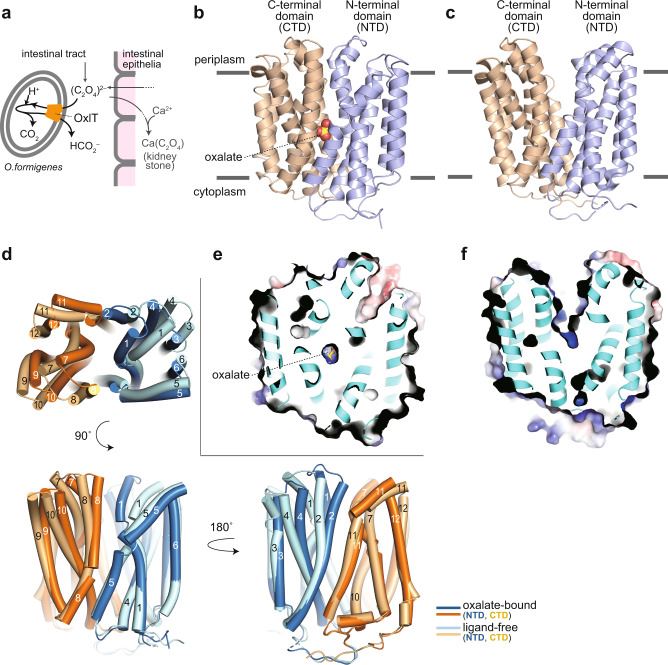 Figure 1. Structure of OxlT. (Jaunet-Lahary T, et al., 2023)
Figure 1. Structure of OxlT. (Jaunet-Lahary T, et al., 2023)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| LacY Lactose Permease Transporter (C154G mutant) (expressed in Cricetulus griseus) | Escherichia coli | X-ray diffraction | 3.60 Å | 1PV7 |
| LacY Lactose Permease (C154G mutant) without substrate at 2 pH values (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.95 Å | 2CFQ |
| LacY Lactose Permease (wild-type) with TDG (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 3.60 Å | 2V8N |
| LacY Lactose Permease with covalently bound MTS-gal (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 3.38 Å | 2Y5Y |
| LacY Lactose Permease Transporter (G46W/G262W mutant) with bound lactose analog (expressed in Escherichia coli) | Escherichia coli str. K-12 substr. DH10B | X-ray diffraction | 3.50 Å | 4OAA |
| LacY Lactose Permease Transporter (G46W, G262W mutant) with bound α-NPG (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 3.31 Å | 4ZYR |
| LacY Lactose Permease Transporter (G46W, G262W mutant) with bound nanobody Nb9039 (expressed in Escherichia coli) | Escherichia coli K-12, Vicugna pacos | X-ray diffraction | 3.30 Å | 5GXB |
| LacY Lactose Permease Transporter (G46W, G262W mutant) with bound nanobody Nb9047 in complex with NPG (expressed in Escherichia coli) | Escherichia coli K-12, Lama glama | X-ray diffraction | 3.00 Å | 6C9W |
| FucP Fucose Transporter in outward-facing conformation (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 3.14 Å | 3O7Q |
| MelB Na+/melibiose symporter (expressed in Escherichia coli) | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | X-ray diffraction | 3.35 Å | 4M64 |
| MelB Na+/melibiose symporter, D59C mutant with bound DDMB (expressed in Escherichia coli) | Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | X-ray diffraction | 3.15 Å | 7L16 |
| XylE proton:xylose symporter with bound D-xylose (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 2.81 Å | 4GBY |
| XylE proton:xylose symporter in partially occluded inward-open state (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 3.80 Å | 4JA3 |
| XylE proton:xylose symporter, inward-facing open conformation (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 3.51 Å | 4QIQ |
| GlcP Glucose/H+ symporter (expressed in Escherichia coli) | Staphylococcus epidermidis ATCC 12228 | X-ray diffraction | 3.20 Å | 4LDS |
| GlpT Glycerol-3-Phosphate Transporter (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 3.30 Å | 1PW4 |
| EmrD Multidrug Transporter (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 3.50 Å | 2GFP |
| PepTSo Oligopeptide-proton symporter (POT family) (expressed in Escherichia coli) | Shewanella oneidensis MR-1 | X-ray diffraction | 3.62 Å | 2XUT |
| PepTSo Oligopeptide-proton symporter (POT family) with bound AlaTyr(Br) (expressed in Escherichia coli) | Shewanella oneidensis MR-1 | X-ray diffraction | 3.15 Å | 4TPH |
| PepTSo Oligopeptide-proton symporter (POT family), inward-open conformation (expressed in Escherichia coli) | Shewanella oneidensis MR-1 | X-ray diffraction | 3.00 Å | 4UVM |
| PepTSo2 Oligopeptide-proton symporter (POT family) (expressed in Escherichia coli) | Shewanella oneidensis MR-1 | Cryo-EM single particle analysis | 4.10 Å | 6JI1 |
| PepTSt Oligopeptide-proton symporter (POT family) (expressed in Escherichia coli) | Streptococcus thermophilus LMG 18311 | X-ray diffraction | 3.30 Å | 4APS |
| PepTSt Oligopeptide-proton symporter (POT family) at 100 K (expressed in Escherichia coli) | Streptococcus thermophilus LMG 18311 | X-ray diffraction | 2.30 Å | 4XNJ |
| PepTSt Oligopeptide-proton symporter (POT family) with bound dipeptide Ala-Leu (expressed in Escherichia coli) | Streptococcus thermophilus LMG 18311 | X-ray diffraction | 2.66 Å | 5OXL |
| PepTSt Oligopeptide-proton symporter (POT family) crystallized in space group P3121 (expressed in Escherichia coli) | Streptococcus thermophilus LMG 18311 | X-ray diffraction | 3.40 Å | 5MMT |
| Proton-dependent oligopeptide transporter (POT) (expressed in Escherichia coli) | Geobacillus kaustophilus | X-ray diffraction | 1.90 Å | 4IKV |
| YbgH peptide transporter (POT family), inward facing conformation (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 3.40 Å | 4Q65 |
| PepT dipeptide transporter (POT family) (expressed in Escherichia coli) | Yersinia enterocolitica subsp. palearctica YE-P4 | X-ray diffraction | 3.02 Å | 4W6V |
| PepTXc mammalian-like peptide transporter (POT family) (expressed in Escherichia coli) | Xanthomonas campestris | X-ray diffraction | 2.10 Å | 6EI3 |
| PepTSh S-Cys-Gly-3M3SH transporter involved in body odor production (expressed in Escherichia coli) | Staphylococcus hominis | X-ray diffraction | 2.50 Å | 6EXS |
| NarU nitrate transporter (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 3.00 Å | 4IU9 |
| NarK nitrate/nitrite exchanger (expressed in Escherichia coli) | Escherichia coli K-12, Mus musculus | X-ray diffraction | 2.60 Å | 4JR9 |
| NarK nitrate/nitrite exchanger, apo inward-open (expressed in Escherichia coli) | Escherichia coli str. K-12 substr. MG1655 | X-ray diffraction | 2.35 Å | 4U4V |
| NRT1.1 nitrate transporter, apo form (expressed in Saccharomyces cerevisiae) | Arabidopsis thaliana | X-ray diffraction | 3.70 Å | 5A2N |
| NRT1.1 nitrate transporter, homodimer in inward-facing conformation (expressed in Spodoptera frugiperda) | Arabidopsis thaliana | X-ray diffraction | 3.25 Å | 4OH3 |
| YajR drug efflux transporter (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 3.15 Å | 3WDO |
| MdfA multidrug resistance transporter in complex with deoxycholate (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 2.00 Å | 4ZP0 |
| MdfA multidrug resistance transporter, E26T/D34M/A150E mutant in the presence of chloramphenicol, pH 5 (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.00 Å | 6VRZ |
| MdfA multidrug resistance transporter, I239T/G354E mutant in complex with LDAO (expressed in Escherichia coli) | Escherichia coli | X-ray diffraction | 2.20 Å | 6OOM |
| MdfA multidrug resistance transporter in outward open conformation (expressed in Escherichia coli, Mus musculus) | Escherichia coli K-12, Mus musculus | X-ray diffraction | 3.40 Å | 6GV1 |
| MdfA multidrug resistance transporter, Q131R/L339E mutant (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 2.20 Å | 6EUQ |
| GLUT1 glucose transporter (N45T/E329Q mutant) (expressed in Trichoplusia ni) | Homo sapiens | X-ray diffraction | 3.17 Å | 4PYP |
| GLUT1 glucose transporter, Wild-type in complex with cytochalasin (expressed in Saccharomyces cerevisiae) | Homo sapiens | X-ray diffraction | 3.00 Å | 5EQI |
| GLUT1 glucose transporter (SLC2A1), inward conformation (expressed in Saccharomyces cerevisiae) | Homo sapiens | X-ray diffraction | 2.40 Å | 6THA |
| GLUT3 glucose transporter (N45T mutant) with bound D-glucose, outward-occluded conformation (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 1.50 Å | 4ZW9 |
| GLUT3 glucose transporter in complex with C3361 (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.30 Å | 7CRZ |
| GLUT3 glucose transporter with bound glucose, exofacial state (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.10 Å | 7SPT |
| GLUT4 glucose transporter with bound cytochalasin B in lipid nanodiscs (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.25 Å | 7WSM |
| GLUT5 fructose transporter, open-inward conformation (expressed in Saccharomyces cerevisiae) | Bos taurus | X-ray diffraction | 3.20 Å | 4YB9 |
| GLUT5 fructose transporter, open-outward conformation (expressed in Saccharomyces cerevisiae, Brevibacillus choshinensis) | Rattus norvegicus, Mus musculus | X-ray diffraction | 3.27 Å | 4YBQ |
| Hexose transporter (HT1) (expressed in Saccharomyces cerevisiae) | Plasmodium falciparum | X-ray diffraction | 3.65 Å | 6RW3 |
| Ferroportin (FPN) Fe2+ transporter (SLC40A1) analog, outward-facing conformation (expressed in Escherichia coli) | Bdellovibrio bacteriovorus HD100 | X-ray diffraction | 2.20 Å | 5AYN |
| Ferroportin (FPN) Fe2+ transporter (SLC40A1), apo protein in nanodisc (expressed in Spodoptera frugiperda, Homo sapiens) | Homo sapiens, Mus musculus | Cryo-EM single particle analysis | 3.20 Å | 6W4S |
| Ferroportin (FPN) Fe2+ transporter (SLC40A1), bound to Ca2+ in nanodisc (expressed in Spodoptera frugiperda) | Homo sapiens, Mus musculus | Cryo-EM single particle analysis | 3.00 Å | 8DL6 |
| Ferroportin (FPN) Fe2+ transporter (SLC40A1) in complex with Fab (expressed in Trichoplusia ni) | Carlito syrichta, Mus musculus | Cryo-EM single particle analysis | 3.00 Å | 6VYH |
| Sugar Transport Protein 10 in complex with glucose in the outward occluded state (expressed in Saccharomyces cerevisiae) | Arabidopsis thaliana | X-ray diffraction | 2.40 Å | 6H7D |
| Sugar Transport Protein 10, outward occluded conformation (expressed in Saccharomyces cerevisiae) | Arabidopsis thaliana | X-ray diffraction | 1.81 Å | 7AAQ |
| LtaA Lipoteichoic acids flippase (expressed in Escherichia coli) | Staphylococcus aureus | X-ray diffraction | 3.30 Å | 6S7V |
| LmrP multidrug transporter, ligand bound outward-open state (expressed in Lactococcus cremoris subsp. cremoris NZ9000) | Lactococcus lactis | X-ray diffraction | 2.90 Å | 6T1Z |
| NupG nucleoside proton symporter (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 3.00 Å | 7DL9 |
| SotB Drug:Proton Antiporter-1 (DHA1), in the inward-occluded conformation (expressed in Escherichia coli) | Escherichia coli K-12 | X-ray diffraction | 3.06 Å | 6KKI |
| MFSD2A lysolipid transporter in complex with LPC-18:3 (expressed in Escherichia coli, Spodoptera frugiperda) | synthetic construct, Gallus gallus | Cryo-EM single particle analysis | 3.03 Å | 7MJS |
| MFSD2A lysolipid transporter (expressed in HEK293 cells) | Mus musculus | Cryo-EM single particle analysis | 3.50 Å | 7N98 |
| MFSD2A lysolipid transporter in complex with syncytin-2 (SYNC2) (expressed in HEK293 cells) | Homo sapiens | Cryo-EM single particle analysis | 3.60 Å | 7OIX |
| OxlT oxalate:formate antiporter (OFA) in complex with fab, oxalate-bound occluded form (expressed in Escherichia coli, Mus musculus) | Oxalobacter formigenes, Mus musculus | X-ray diffraction | 3.00 Å | 8HPK |
Table 1. Structural research of MFS transporters.
Creative Biostructure stands as a prominent organization in the realm of structural biology, occupying a leading position in providing structural analysis services for crucial membrane proteins. Leveraging our advanced cryo-electron microscopy (cryo-EM), X-ray crystallography, and NMR spectroscopy technologies, we have achieved exceptional success in unraveling the intricate structures of diverse transporters at high resolutions. Through our contributions, we have significantly advanced the understanding of their functional mechanisms. Contact us today and unlock the potential of our advanced capabilities to empower your research, propelling you closer to realizing your scientific objectives.
References
- Huang J, et al. Orthosteric–allosteric dual inhibitors of PfHT1 as selective antimalarial agents. Proceedings of the National Academy of Sciences. 2021, 118(3): e2017749118.
- Wang N, et al. Molecular basis for inhibiting human glucose transporters by exofacial inhibitors. Nature Communications. 2022, 13(1): 2632.
- Shen J, et al. Mechanism of Ca2+ transport by ferroportin. Elife. 2023, 12: e82947.
- Jaunet-Lahary T, et al. Structure and mechanism of oxalate transporter OxlT in an oxalate-degrading bacterium in the gut microbiota. Nature Communications. 2023, 14(1): 1730.