Structural Research of Nitric Oxide Reductases
Nitric oxide reductase (NOR) is an iron-containing membrane integrase. It catalyzes 2e− reduction of nitric oxide (NO) to nitrous oxide (N2O) and plays an important role in denitrification, NO detoxification and NO-mediated intercellular signaling. Based on the amino acid sequence of the core part of NOR, it is classified as a heme-copper oxidase (HCO) superfamily. Three types of NOR have been identified from bacteria: cNOR, qNOR, and qCuNOR.
Study on the Structure of Pseudomonas aeruginosa cNOR and thermogeobacillus lipophilus qNOR
Both cNOR and qNOR TM contain heme b, heme b3 and non-heme metals as active centers. In cNOR, the cytochrome c fold is typical. The NorB subunit contains 12 TM helices, and the NorC subunit has an α-helix in the TM region and a hydrophilic domain on the periplasmic side. qNOR contains 14 TM helices in the transmembrane region and an alpha-helical hydrophilic domain outside the cell. The helical topologies of TM regions in the two NORs are basically similar. NOR has five conserved Glu residues that play a core role in catalyzing NO reduction.
Molecular Mechanism of cNOR and qNOR
When NO is reduced by cNOR, two NO molecules are accommodated in the binuclear centers of heme b3 and FeB. The change in the amino acid ligand causes the two NO molecules to form an arrangement suitable for the N\N bond in the center of the double nucleus. The Glu ligand of qNOR can briefly separate from FeB, thereby expanding the space in the binuclear center to accommodate a second NO molecule. In cNOR and qNOR, there is no continuous channel or hydrogen bond network between the cytoplasm and the extracellular/periplasmic side, resulting in protons not permeating through the TM region of NORs.
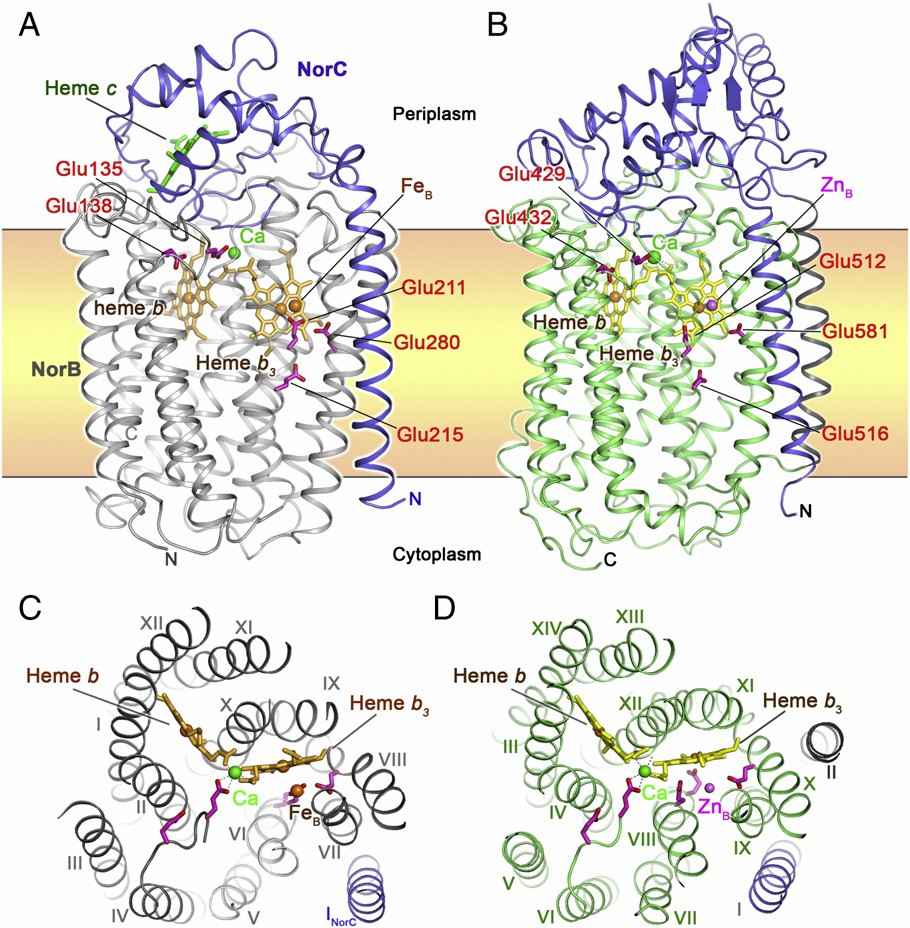 Figure 1. The overall structure and top view of cNOR and qNOR. (Shiro Y., 2012)
Figure 1. The overall structure and top view of cNOR and qNOR. (Shiro Y., 2012)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Dimeric quinol dependent nitric oxide reductase (qNOR) | Achromobacter xylosoxidans | Cryo-EM single particle analysis | 3.9 Å | 6QQ5 |
| Nitric oxide reductase | Roseobacter denitrificans OCh 114 | X-ray diffraction | 2.85 Å | 4XYD |
| Nitric oxide reductase | Geobacillus stearothermophilus | X-ray diffraction | 2.5 Å | 3AYF |
| nitric oxide reductase complex with HQNO | Geobacillus stearothermophilus | X-ray diffraction | 2.7 Å | 3AYG |
| Glu-494-Ala inactive monomer of a quinol dependent Nitric Oxide Reductase (qNOR) | Achromobacter xylosoxidans | Cryo-EM single particle analysis | 4.50 Å | 6T6V |
| The complex of Cytochrome cd1 Nitrite Reductase and Nitric Oxide Reductase in Denitrification | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 3.2 Å | 5GUW |
| nitric oxide reductase in complex with antibody fragment | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 2.7 Å | 3O0R |
| Reduced cytochrome c-dependent nitric oxide reductase (cNOR) in complex with antibody fragment | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 2.7 Å | 2WFB |
| Cytochrome c-dependent nitric oxide reductase (cNOR) in complex with xenon | Pseudomonas aeruginosa PAO1 | X-ray diffraction | 3.3 Å | 5GUX |
| No complex of thr243ala mutants of cytochrome p450nor | Fusarium oxysporum | X-ray diffraction | 1.4 Å | 1F24 |
| Oxidized (di-ferric) FprA | Moorella thermoacetica | X-ray diffraction | 3 Å | 1YCF |
| Neisseria meningitidis qNOR | Neisseria meningitidis alpha14 | X-ray diffraction | 4.2 Å | 6FWF |
| Quinol-dependent nitric oxide reductase (qNOR) in the monomeric oxidized state with zinc complex. | Neisseria meningitidis alpha14 | X-ray diffraction | 3.15 Å | 6L1X |
| Flavodiiron protein D52K mutant in the oxidized state | Escherichia coli K-12 | X-ray diffraction | 1.978 Å | 7ROF |
Table 1. Structural research of nitric oxide reductases.
To study the structure of membrane proteins, cryo-electron microscopy (cryo-EM) and X-ray crystallography are commonly used. Structural analysis of nitric oxide reductases is beneficial to the study of biomolecular functions. It also helps in the development of new targeted drugs.
Creative Biostructure has long been committed to the study of structural biology and membrane proteins. We have extensive experience in determining membrane protein structures using single particle analysis (SPA). In addition to the structural determination of membrane proteins, we can accurately analyze biomolecules, including but not limited to nucleic acids, ribosomes, small proteins, protein complexes, protein-ligand complexes, and viruses. If you are interested in our services, please contact us and we will provide you with a professional and comprehensive solution.
References
- Shiro Y. Structure and function of bacterial nitric oxide reductases: nitric oxide reductase, anaerobic enzymes. Biochim Biophys Acta. 2012;1817(10):1907-1913.
- Collman JP, et al. A functional nitric oxide reductase model. Proc Natl Acad Sci U S A. 2008;105(41):15660-15665.