Structural Research of Photoprotection Proteins
Photoprotective proteins help protect organisms from the harmful effects of excessive light exposure, particularly in the photosynthetic system, and are involved in dissipating excess energy, preventing photo-oxidative damage, and maintaining optimal photosynthetic efficiency. Common photoprotective proteins include light-harvesting complexes (LHC), non-photochemical burst (NPQ) proteins, and photolytic enzymes. Exploring the structures of photoprotective proteins can reveal the action mechanisms in preventing photoinhibition and oxidative stress.
Structural analysis of orange carotenoid protein (OCP)
Orange carotenoid protein (OCP) is a typical photoprotective protein that controls photoprotection in many cyanobacteria. It is structurally modular and consists of the N-terminal domain NTD (an all-alpha helix consisting of two staggered 4-helix bundles) and the C-terminal domain CTD (mixed α/β). These two structural domains are connected by an extended linker that carotenoids span during. Further studies revealed that NTD is responsible for the non-photochemical bursting mechanism of OCP.
Research on the structure of photoprotective protein PsbS
Recent advances in the field of single-particle cryo-electron microscopy have allowed the structure of photoprotective proteins to be elucidated and analyzed. PsbS, for example, is a photoprotective protein to ensures the tolerance of the plant photosynthetic apparatus (PSA) to light changes. The low pH crystal structure of PsbS shows that it has four transmembrane α-helices, TM1, 2, 3, and 4, connected by a relatively short portion of the lumen, and a short α-helix between TM3 and TM4 that is roughly parallel to the membrane. TM2 and TM3 are connected by a large matrix-exposed loop.
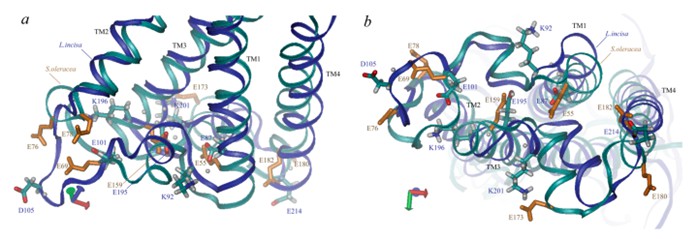 Figure 1. Three-dimensional structural view of PsbS along the membrane (a) and lumen side (b). (Ptushenko VV, et al., 2023)
Figure 1. Three-dimensional structural view of PsbS along the membrane (a) and lumen side (b). (Ptushenko VV, et al., 2023)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| The photoprotective protein PsbS | Spinacia oleracea | X-ray diffraction | 2.35 Å | 4RI2 |
| DCCD-modified PsbS | Spinacia oleracea | X-ray diffraction | 2.7 Å | 4RI3 |
| Fluorescence recovery-like protein FRPL | Pseudomonas borbori | X-ray diffraction | 1.8 Å | 8AG8 |
| Fluorescence recovery protein | Synechocystis sp. PCC 6803 substr. Kazusa | X-ray diffraction | 2.5 Å | 4JDX |
| The N-terminal domain of OCP binding canthaxanthin | Synechocystis sp. PCC 6803 substr. Kazusa | X-ray diffraction | 1.544 Å | 4XB4 |
| Orange carotenoid protein binding canthaxanthin | Synechocystis sp. PCC 6803 substr. Kazusa | X-ray diffraction | 1.9 Å | 4XB5 |
| Orange carotenoid binding protein 1 (OCP1) | Gloeocapsa sp. PCC 7513 | X-ray diffraction | 2.17 Å | 7EKR |
| Orange carotenoid binding protein 1 (OCP1) | Tolypothrix sp. PCC 7601 | X-ray diffraction | 1.61 Å | 6PQ1 |
| Orange carotenoid protein | Synechocystis sp. PCC 6803 | X-ray diffraction | 1.649 Å | 3MG1 |
| Orange carotenoid protein Y44S mutant | Synechocystis sp. PCC 6803 | X-ray diffraction | 2.653 Å | 3MG2 |
| Orange carotenoid protein R155L mutant | Synechocystis sp. PCC 6803 | X-ray diffraction | 1.702 Å | 3MG3 |
| PEA light-harvesting complex II | Pisum sativum | X-ray diffraction | 2.5 Å | 2BHW |
| Orange carotenoid protein binding canthaxanthin in the C2 space group | Planktothrix agardhii | X-ray diffraction | 1.85 Å | 7QCZ |
| Orange carotenoid protein binding echinenone in the P21 space group | Planktothrix agardhii | X-ray diffraction | 1.71 Å | 7QD1 |
| Water-soluble chlorophyll protein | Lepidium virginicum | X-ray diffraction | 2 Å | 2DER |
| Bry-LHCII homotrimer | Bryopsis corticulans | Cryo-EM single particle analysis | 2.55 Å | 8HLV |
| Bry-LHCII heterotrimer | Bryopsis corticulans | Cryo-EM single particle analysis | 2.74 Å | 8HPD |
Table 1. Structural research of photoprotection proteins.
As a pioneer in the field of structural biology, Creative Biostructure provides cutting-edge structural analysis services. Our expertise in NMR spectroscopy, cryo-electron microscopy (cryo-EM) and X-ray crystallography allows us to determine the high-resolution structure of photoprotection proteins. We utilize advanced technology and an in-depth understanding of structural biology to provide comprehensive insights into the molecular mechanisms of various biological processes.
If you need to ensure accurate and comprehensive analysis and understanding of protein structure and function, contact us to learn how our cutting-edge capabilities can enhance your research and propel you closer to achieving your scientific goals.
References
- Ptushenko VV, et al. The Photoprotective Protein PsbS from Green Microalga Lobosphaera incisa: The Amino Acid Sequence, 3D Structure and Probable pH-Sensitive Residues. International Journal of Molecular Sciences. 2023. 24(20):15060.
- Fan M, et al. Crystal structures of the PsbS protein essential for photoprotection in plants. Nat Struct Mol Biol. 2015. 22(9):729-735.
- Kerfeld CA, et al. The crystal structure of a cyanobacterial water-soluble carotenoid binding protein. Structure. 2003. 11(1):55-65.