Structural Research of G Protein-coupled Receptors (GPCRs) Class A
G protein-coupled receptors (GPCRs) are integral membrane proteins that play essential roles in cellular signaling and communication. Among the GPCRs, class A receptors are the largest and most extensively studied subgroup, with significant implications in various physiological processes, including neurotransmission, hormone regulation, and immune responses. In recent decades, structural research on class A GPCRs has significantly advanced, shedding light on their activation mechanisms and providing insights into the development of novel therapeutics.
Class A GPCRs are characterized by their seven-transmembrane helices and their activation through G protein coupling. These receptors are of great interest to researchers due to their involvement in a wide range of physiological functions and their potential as drug targets. Early research on GPCRs mainly relied on X-ray crystallography, which led to landmark discoveries, such as the first high-resolution structure of a GPCR, rhodopsin, in 2000.
Over the past two decades, the advent of cryo-electron microscopy (cryo-EM) has revolutionized structural biology, allowing the visualization of GPCRs at near-atomic resolution. The latest cryo-EM structures of GPR119 (a cannabinoid receptor-like class A GPCR) and its interactions with agonists provide significant revelations regarding its activation and signaling mechanisms, thereby contributing to the advancement of innovative therapeutic strategies.
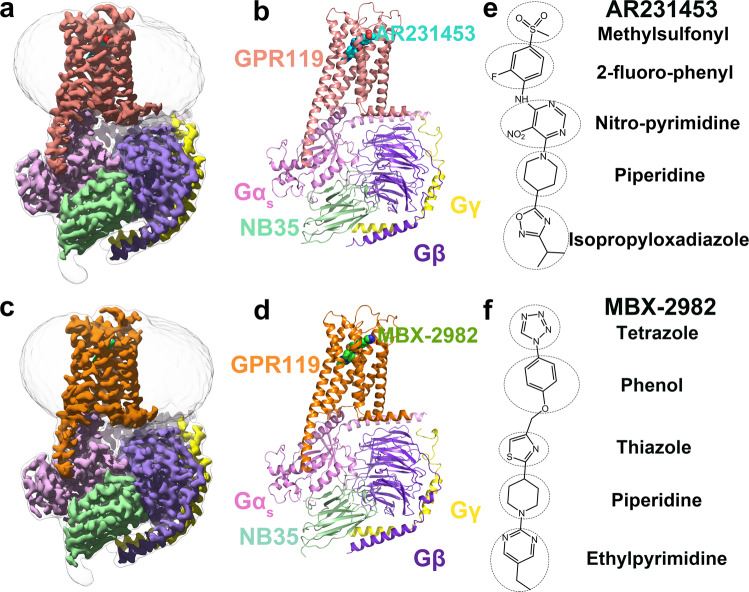 Figure 1. Overall cryo-EM structures of the GPR119-Gs heterotrimer complexes. (Qian Y, et al., 2022)
Figure 1. Overall cryo-EM structures of the GPR119-Gs heterotrimer complexes. (Qian Y, et al., 2022)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Rhodopsin in ligand-free state (opsin) | Bos taurus | X-ray diffraction | 2.65 Å | 4J4Q |
| Rhodopsin in ligand-free state (opsin) | Bos taurus | X-ray diffraction | 2.29 Å | 4X1H |
| Rhodopsin in ligand-free state (opsin) in complex with ArrFL-1 | Bos taurus | X-ray diffraction | 2.75 Å | 4PXF |
| Rhodopsin, N2C/D282C stabilized opsin bound to RS01 (expressed in HEK293S cells) | Bos taurus | X-ray diffraction | 2.36 Å | 6FK6 |
| Rhodopsin-Gαi-βγ complex (expressed in HEK293 cells) | Bos taurus | Cryo-EM single particle analysis | 4.38 Å | 6QNO |
| Rhodopsin-Transducin Complex (expressed in Escherichia coli) | Bos taurus | Cryo-EM single particle analysis | 3.90 Å | 6OY9 |
| Visual Signaling Complex between Transducin and Phosphodiesterase 6 (expressed in Escherichia coli) | Bos taurus | Cryo-EM single particle analysis | 3.20 Å | 7JSN |
| Rhodopsin (native dimer) in nanodiscs | Bos taurus | Cryo-EM single particle analysis | 4.50 Å | 6OFJ |
| Rhodopsin in complex with rhodopsin kinase (GRK1) (expressed in Spodoptera frugiperda) | Bos taurus | Cryo-EM single particle analysis | 7.00 Å | 7MT9 |
| Bovine rhodopsin in Lipidic Cubic Phase (SACLA) | Bos taurus | X-ray diffraction | 1.80 Å | 7ZBC |
| Human rhodopsin with bound mouse visual arrestin (expressed in HEK293S cells) | Homo sapiens | X-ray diffraction | 3.30 Å | 4ZWJ |
| Human rhodopsin with bound inhibitory G protein (Gi) (expressed in Sf9 cells) | Homo sapiens | Cryo-EM single particle analysis | 4.50 Å | 6CMO |
| Jumping Spider Rhodopsin-1 bound to 9-cis retinal (expressed in HEK293 cells) | Hasarius adansoni | X-ray diffraction | 2.15 Å | 6I9K |
| GPR17-Gi complex (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.02 Å | 7Y89 |
| Gi bound orphan GPR20 in ligand-free state (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.14 Å | 8HS3 |
| GPR21-Gs complex (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 2.91 Å | 8HMV |
| GPR52 ligand free form with flavodoxin fusion (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.90 Å | 6LI1 |
| Orphan GPR88 class A receptor - Gi1 complex, apo form (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.00 Å | 7WZ4 |
| GPR119-Gs-LPC complex (expressed in Trichoplusia ni) | Homo sapiens | Cryo-EM single particle analysis | 3.10 Å | 7XZ5 |
| GPR119-Gs Complex with small molecule agonist AR231453 (expressed in Trichoplusia ni) | Homo sapiens | Cryo-EM single particle analysis | 2.87 Å | 7WCN |
| Orphan GPCR in complex with Gi (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.20 Å | 7VUG |
| LPS-bound GPR174 in complex with Gs protein (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 2.76 Å | 7XV3 |
| GSK682753A-bound EBI2/GPR183 (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 2.98 Å | 7TUY |
| OX1 orexin receptor with bound suvorexant (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.75 Å | 4ZJ8 |
| Orexin-1 receptor in complex with suvorexant (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.26 Å | 6TO7 |
| OX1 orexin receptor with bound subtype-selective antagonist (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 3.50 Å | 6V9S |
| OX2 orexin receptor with bound suvorexant (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.50 Å | 4S0V |
| OX2 orexin receptor in complex with suvorexant (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.74 Å | 6TPJ |
| OX2 orexin receptor with bound antagonist EMPA (expressed in Sf9 cells) | Homo sapiens | X-ray diffraction | 1.96 Å | 5WQC |
| OX2 orexin receptor in complex with G protein and natural peptide-agonist Orexin B (OxB) (expressed in Trichoplusia ni) | Homo sapiens | Cryo-EM single particle analysis | 3.20 Å | 7L1U |
| OX2 orexin receptor in complex with Gi protein and TAK-925 (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.17 Å | 7SQO |
| OX2 orexin receptor with bound lemborexant (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.89 Å | 7XRR |
| C5a anaphylatoxin chemotactic receptor 1 (C5aR) in complex with NDT9513727 (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.70 Å | 5O9H |
| C5a anaphylatoxin chemotactic receptor 1 (C5aR) in complex with orthosteric antagonist PMX53 and allosteric antagonist NDT9513727 (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.90 Å | 6C1Q |
| α1B-adrenergic receptor in complex with inverse agonist (+)-cyclazosin (expressed in Escherichia coli) | Homo sapiens | X-ray diffraction | 2.87 Å | 7B6W |
| alpha2A adrenergic receptor in complex with an antagonist RSC (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.70 Å | 6KUX |
| alpha2A adrenergic receptor GoA signaling complex bound to a G protein biased agonist (expressed in Spodoptera frugiperda) | Homo sapiens | Cryo-EM single particle analysis | 3.47 Å | 7W6P |
| alpha2BAR-GoA complex (expressed in Sf9 cells) | Homo sapiens | Cryo-EM single particle analysis | 2.90 Å | 6K41 |
| alpha2C adrenergic G protein-coupled receptor (expressed in Spodoptera frugiperda) | Homo sapiens | X-ray diffraction | 2.80 Å | 6KUW |
Table 1. Structural research of class-A GPCRs.
Creative Biostructure offers comprehensive structural analysis services for class A GPCRs, tailored to meet the specific needs of researchers and pharmaceutical companies. Our multidisciplinary approach combines cryo-electron microscopy (cryo-EM), X-ray crystallography, and computational modeling to provide a holistic understanding of GPCR structures and their interactions with ligands and G proteins.
Researchers and pharmaceutical companies seeking to gain a deeper understanding of GPCRs and harness this knowledge to develop novel therapies are encouraged to collaborate with Creative Biostructure. Our state-of-the-art facilities, experienced team, and track record of successful structural studies make us a reliable partner in advancing the field of GPCR research. Contact us to discover how our cutting-edge capabilities can empower your research and drive you closer to achieving your scientific goals.
References
- Chen G, et al. Activation and allosteric regulation of the orphan GPR88-Gi1 signaling complex. Nature Communications. 2022, 13(1): 2375.
- Qian Y, et al. Activation and signaling mechanism revealed by GPR119-Gs complex structures. Nature Communications. 2022, 13(1): 7033.