Structural Research of SNARE Protein Family
Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) are a family of small conserved eukaryotic proteins that mediate membrane fusion between organelles and plasma membranes. The most well-researched are SNAREs that mediate the release of neurotransmitter-containing synaptic vesicles in neurons. These neuronal traps are targets of neurotoxins produced by certain bacteria that cause botulism and tetanus.
Structural analysis of SNARE
SNAREs play essential roles in vesicular transport and are usually inserted post-translationally into membranes via C-terminal transmembrane structural domains, including insertion into the plasma membrane, endoplasmic reticulum, mitochondria, peroxisomes, etc. SNAREs are targeted by altering the composition of the amino acid residues flanking the C-terminus or the length of the transmembrane structural domains. Although SNAREs vary greatly in structure and size, they all contain a SNARE motif of 60-70 amino acids in their cytoplasmic domains and heptameric repeats capable of forming convoluted helices. v- and t-SNAREs can be reversibly assembled into tightly packed four-helix "trans"-SNARE complexes. SNARE complexes.
Structural resolution of synaptotagmin-1 and neuronal SNARE complexes
Using diffraction data from an X-ray free-electron laser, researchers have determined atomic-resolution structures of synaptotagmin-1 and neuronal SNARE complexes with precise side-chain spindle assignments. The crystal structure of Syt1-SNARE reveals three interfaces between the SNARE complex and the Syt1 C2A and C2B structural domains. In the long-cell form, two instances of essentially identical primary interfaces are formed between the Syt1 C2B structural domain and the SNARE complex. The secondary interface involves another Syt1 C2B structural domain and the SNARE complex, while the tertiary interface involves the Syt1 C2A structural domain and the SNARE complex. The C2A structural domains forming the tertiary interface have the same orientation in both crystal forms and the interacting side chains are located in similar positions.
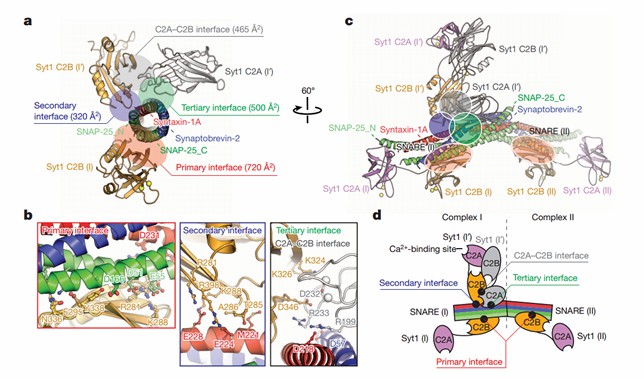 Figure 1. Crystal structure of the Syt1–SNARE complex. (Zhou Q, et al., 2015)
Figure 1. Crystal structure of the Syt1–SNARE complex. (Zhou Q, et al., 2015)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| SM Protein Vps45 in Complex with Qa SNARE Tlg2 (1-310) | Thermochaetoides thermophila | X-ray diffraction | 3.9 Å | 6XMD |
| SM Protein Vps45 in Complex with Qa SNARE Tlg2 | Thermochaetoides thermophila | X-ray diffraction | 2.8 Å | 6XM1 |
| A complex between the SNARE Vam3 and the HOPS Vps33-Vps16 subcomplex | Thermochaetoides thermophila DSM 1495 | X-ray diffraction | 3.1 Å | 5BUZ |
| A complex between the SNARE Nyv1 and the HOPS Vps33-Vps16 subcomplex | Thermochaetoides thermophila DSM 1495 | X-ray diffraction | 3.1 Å | 5BV0 |
| Vps33-Vps16 Complex | Thermochaetoides thermophila DSM 1495 | X-ray diffraction | 2.902 Å | 5BV1 |
| Syntaxin 6 | Rattus norvegicus | X-ray diffraction | 2.1 Å | 1LVF |
| SM protein Vps45 | Thermochaetoides thermophila | X-ray diffraction | 2 Å | 6XJL |
| The vesicular transport protein Sec17 | Saccharomyces cerevisiae | X-ray diffraction | 2.9 Å | 1QQE |
| NSF-D1D2 part in the whole 20S complex | Cricetulus griseus | Cryo-EM single particle analysis | 3.7 Å | 6IP2 |
| STX17 LIR region in complex with GABARAP | Homo sapiens | X-ray diffraction | 2 Å | 7BV4 |
| ATP-bound N-ethylmaleimide sensitive factor | Cricetulus griseus | Cryo-EM single particle analysis | 4.2 Å | 3J94 |
| The neuronal complexin/snare complex | Doryteuthis pealeii | X-ray diffraction | 2.95 Å | 1L4A |
| Autophagic STX17/SNAP29/VAMP8 SNARE complex | Homo sapiens | X-ray diffraction | 3.05 Å | 7BV6 |
| Unc18-syntaxin 1 complex | Monosiga brevicollis | X-ray diffraction | 2.8 Å | 2XHE |
| HOPS component Vps33 | Thermochaetoides thermophila DSM 1495 | X-ray diffraction | 2.6 Å | 4JC8 |
| Gamma-SNAP | Danio rerio | X-ray diffraction | 2.6 Å | 2IFU |
| Yeast T-SNARE protein SSO1 | Saccharomyces cerevisiae | X-ray diffraction | 2.1 Å | 1FIO |
| SNARE Sec20 bound to Dsl1 complex subunit Tip20 | Eremothecium gossypii ATCC 10895 | X-ray diffraction | 3.203 Å | 6WC3 |
| Sly1 N-terminal domain | Rattus norvegicus | SOLUTION NMR | / | 1Y9J |
| Sro7 | Saccharomyces cerevisiae | X-ray diffraction | 2.4 Å | 2OAJ |
| Lipid-bound synaptobrevin | Rattus norvegicus | SOLUTION NMR | / | 2KOG |
| Sly1p in complex with an N-terminal peptide of Sed5p | Saccharomyces cerevisiae | X-ray diffraction | 3 Å | 1MQS |
| Sec22b N-terminal domain | Mus musculus | X-ray diffraction | 2.4 Å | 1IFQ |
| Neuronal SNARE Syntaxin 1A | Rattus norvegicus | SOLUTION NMR | / | 2M8R |
| Neuronal SEC1 | Doryteuthis pealeii | X-ray diffraction | 2.4 Å | 1EPU |
| ADP-bound N-ethylmaleimide sensitive factor | Cricetulus griseus | Cryo-EM single particle analysis | 7.6 Å | 3J95 |
| GppNHp-Bound Ypt1p GTPase | Saccharomyces cerevisiae | X-ray diffraction | 2.06 Å | 1YZN |
| Mint1/Munc18-1/syntaxin-1 complex | Rattus norvegicus | X-ray diffraction | 3.2 Å | 7XSJ |
Table 1. Structural research of the SNARE protein family.
Creative Biostructure offers a range of techniques for determining the three-dimensional structure of proteins, such as X-ray crystallography, cryo-electron microscopy (cryo-EM), and nuclear magnetic resonance (NMR), which help researchers determine the atomic structure of complex membrane proteins. Our services enable structural analysis of SNARE protein family members and provide valuable insights into their action mechanisms in the nervous system.
As a leader in protein structural analysis services, we are committed to advancing structural biology research and providing high-quality, comprehensive services to our clients. If you are interested in our services, please contact us today to learn more details.
References
- Zhou Q, et al. Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature. 2015. 525(7567): 62-67.
- Wang T, et al. SNARE proteins in membrane trafficking. Traffic. 2017.18(12): 767-775.
- Han J, et al. The Multifaceted Role of SNARE Proteins in Membrane Fusion. Front Physiol. 2017. 8: 5.
- Fasshauer D. Structural insights into the SNARE mechanism. Biochim Biophys Acta. 2003.1641(2-3): 87-97.