Structural Research of Outer Membrane Autotransporters
Autotransporter protein (AT) is a core component of the outer membrane of Gram-negative bacteria that transports biomolecules with the help of β-barrel transmembrane structural domains. AT plays a central role in controlling bacterial interactions with the host or the environment as a part of the V-secretory system. It promotes biofilm formation, surface adhesion, host invasion, cytotoxicity, and immunomodulation. Studying the structure of AT is beneficial for revealing its cytotoxic function and further elucidating the pathogenesis of bacteria.
The structure and action mechanism of type 5 secretory system (T5SS)
T5SS is the largest secretory protein group in Gram-negative bacteria, divided into Va to Vf subtypes, with each protein having a secretory function. In the cytoplasm, AT carries an N-terminal signaling peptide (SP), which is used for Sec-mediated transmembrane transport. The periplasmic partner keeps the AT in an unfolded state and reaches the outer membrane. The translocator forms a pore in the outer membrane, providing the driving force through protein folding to transport biological molecules to the cell surface.
Research progress on the structure of autotransporter Ssp
Ssp comes from Serratia marcescens. Determine the three-dimensional structure of Ssp through X-ray diffraction. Obtain crystals of unlabeled and SeMet labeled Ssp, and use the homologous model server SWISS-MODEL to establish a preliminary model of Ssp based on the IcsA automatic transporter (PDB: 5KE1). Merge the SeMet dataset and use SIRAS and MRSAD to solve the structure. The structure of Ssp indicates that it consists of an N-terminal Bacillus subtilis protease-like protease structural domain and a C-terminal triple-stranded β-helical structural domain.
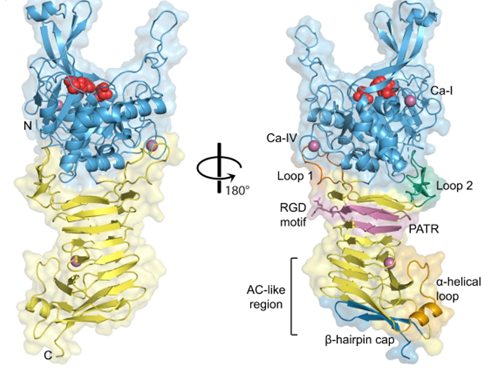 Figure 1. Crystal structure of Ssp. (Hor L, et al., 2023)
Figure 1. Crystal structure of Ssp. (Hor L, et al., 2023)
| Protein | Organism | Method | Resolution | PDB Entry ID |
| Translocator domain of autotransporter NalP | Neisseria meningitidis | X-ray diffraction | 3.2 Å | 1UYO |
| Translocator domain of autotransporter NalP | Neisseria meningitidis | X-ray diffraction | 2.6 Å | 1UYN |
| EspP autotransporter Beta-domain. | Escherichia coli O157:H7 | X-ray diffraction | 2.66 Å | 2QOM |
| Autotransporter EspP - N1023A mutant | Escherichia coli O157:H7 | X-ray diffraction | 2.481 Å | 3SLJ |
| Beta Domain of the Autotransporter Brka | Bordetella pertussis | X-ray diffraction | 3 Å | 3QQ2 |
| Transport unit of the autotransporter AIDA-I | Escherichia coli | X-ray diffraction | 3 Å | 4MEE |
| Integral membrane domain of autotransporter Hbp | Escherichia coli | X-ray diffraction | 2 Å | 3AEH |
| Full-length autotransporter EstA | Pseudomonas aeruginosa | X-ray diffraction | 2.499 Å | 3KVN |
| The passenger domain of autotransporter EspP | Escherichia coli O157:H7 str. EDL933 | X-ray diffraction | 2.5 Å | 3SZE |
| TibC-catalyzed hyper-glycosylated TibA55-350 fragment | Escherichia coli ETEC H10407 | X-ray diffraction | 2.11 Å | 4Q1Q |
| Hap adhesin | Haemophilus influenzae | X-ray diffraction | 2.2 Å | 3SYJ |
| Hia 1022-1098 | Haemophilus influenzae | X-ray diffraction | 2 Å | 2GR8 |
| Hia 992-1098 | Haemophilus influenzae | X-ray diffraction | 2.3 Å | 2GR7 |
| A C-terminal fragment of the IcsA/VirG passenger-domain | Shigella flexneri | X-ray diffraction | 1.9 Å | 5KE1 |
| Autotransporter EspP - N1023D mutant | Escherichia coli O157:H7 | X-ray diffraction | 2.52 Å | 3SLO |
| The IcsA autochaperone region | Shigella flexneri | X-ray diffraction | 2 Å | 3LM3 |
| The autotransporter UpaB strain CFT073 | Escherichia coli CFT073 | X-ray diffraction | 1.97 Å | 6BEA |
| The membrane anchor domain of the trimeric autotransporter YadA | Yersinia enterocolitica subsp. enterocolitica 8081 | SOLID-STATE NMR | / | 2LME |
| The LysM region of the E. coli Intimin periplasmic domain | Escherichia coli | SOLID-STATE NMR | / | 2MPW |
Table 1. Structural research of outer membrane autotransporters.
Creative Biostructure provides NMR spectroscopy, X-ray crystallography and cryo-electron microscopy (cryo-EM) to assist clients in studying outer membrane autotransporter structures, to further understand the transport mechanism of bacterial outer membranes, which will provide ideas for the development of new drugs.
We have long been committed to the study of structural biology and membrane proteins. Our experts have extensive experience in determining membrane protein structures. If you are interested in our services, please contact us for more details.
References
- Hor L, et al. Crystal structure of a subtilisin-like autotransporter passenger domain reveals insights into its cytotoxic function. Nat Commun. 2023.14(1):1163.
- Clarke KR, et al. Phylogenetic Classification and Functional Review of Autotransporters. Front Immunol. 2022.13:921272.