De novo Design
De novo design is an attractive approach for constructing designed proteins with predetermined structures and functions. Creative Biostructure provides de novo design related services for protein engineering, including protein structure prediction, protein folding and interactions, thermodynamics information, energetic state, stability, etc.
De novo design requires deep understanding of the structure of a protein and the correlated functions. It is accomplished either from scratch or by the modular approach.
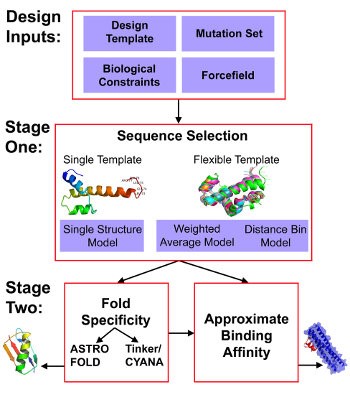 Figure 1. General workflow of De Novo Design strategy
Figure 1. General workflow of De Novo Design strategy
Linear chains of amino acids are adopted a unique 3D structure in the native surroundings to perform proteins. In de novo design, to identify amino acid sequences folding into proteins with desired functions is the ultimate objective. There are various approaches predicting protein structure including comparative modeling and fold recognition. In proteins, sequence similarity implies structural similarity. Accordingly, the method of comparative modeling is formed. The structure of a protein can be predicted by comparing the amino acid sequence to that of native 3D structure known. When the target and template share over 50% of sequence, the predictions are of high quality. Sequence similarity implies structural similarity but similar structures can be found for proteins with different sequence.
As a complemental method, fold recognition aims at predicting the three-dimensional folded structure of a protein with known sequence. The structure is evolutionary more conserved than sequence. As a result, the repertoire of different folds is more limited. The methods of fold recognition mainly include advances sequence comparison and secondary structure prediction and comparison. Besides these, the prediction of loop structure including β-strands and helices is also included.
The proteins designed by de novo which is consistent with the natural 3D targets with minimum energy interactions often can fold very fast. De novo protein design can promote stability of the target protein and also has been used to lock proteins into certain useful conformations. There have been numerous successes in the development of computational algorithms for protein design.
Common approaches used by de novo design include:
- Structural motif: such as “four α-helix bundle” motif, “helix-loop-helix” motif
- Known protein structures as natural scaffolds
- Molecular templates
Popular strategies for protein engineering:
- Rational design
- De novo design
- Directed evolution
- Semi-rational design
Please feel free to contact us for a detailed quote.
Ordering Process
Reference:
C.A. Floudas, et al. Advances in protein structure prediction and de novo protein design: A review. Chemical Engineering Science 61 (2006) 966 – 988.

