Site-saturation Mutagenesis Libraries
Creative Biostructure is highly-experienced in custom mutagenesis libraries construction services for directed evolution. Site-saturation mutagenesis (SSM) can generate random or targeted mutations and screen the library of protein variants for functional alterations. Notably, our protein evolution and engineering platform holds many advantages for custom SSM libraries services, including extremely cost-efficient, fast delivery time, fewer variants to screen and, systematic identification of beneficial amino acid substitutions.
Site-saturation mutagenesis (SSM) is a simple, fast and efficient technique using PCR amplication with degenerate synthetic oligonucleotides as primers. Since a specific amino acid residue within the protein sequence can be identified to be critical for the protein’s function, it is very important to determine the ideal amino acid residue for this position. SSM is a form of random mutagenesis, allowing the substitution of specific sites against all 20 possible amino acids at once. It is well-known as sequential permutation, SSM used by Creative Biostructure in combination with the powerful screeing assay is the most systematic mutagenesis strategy to identify amino acid substitutions that fulfill customers’ protein engineering projects.
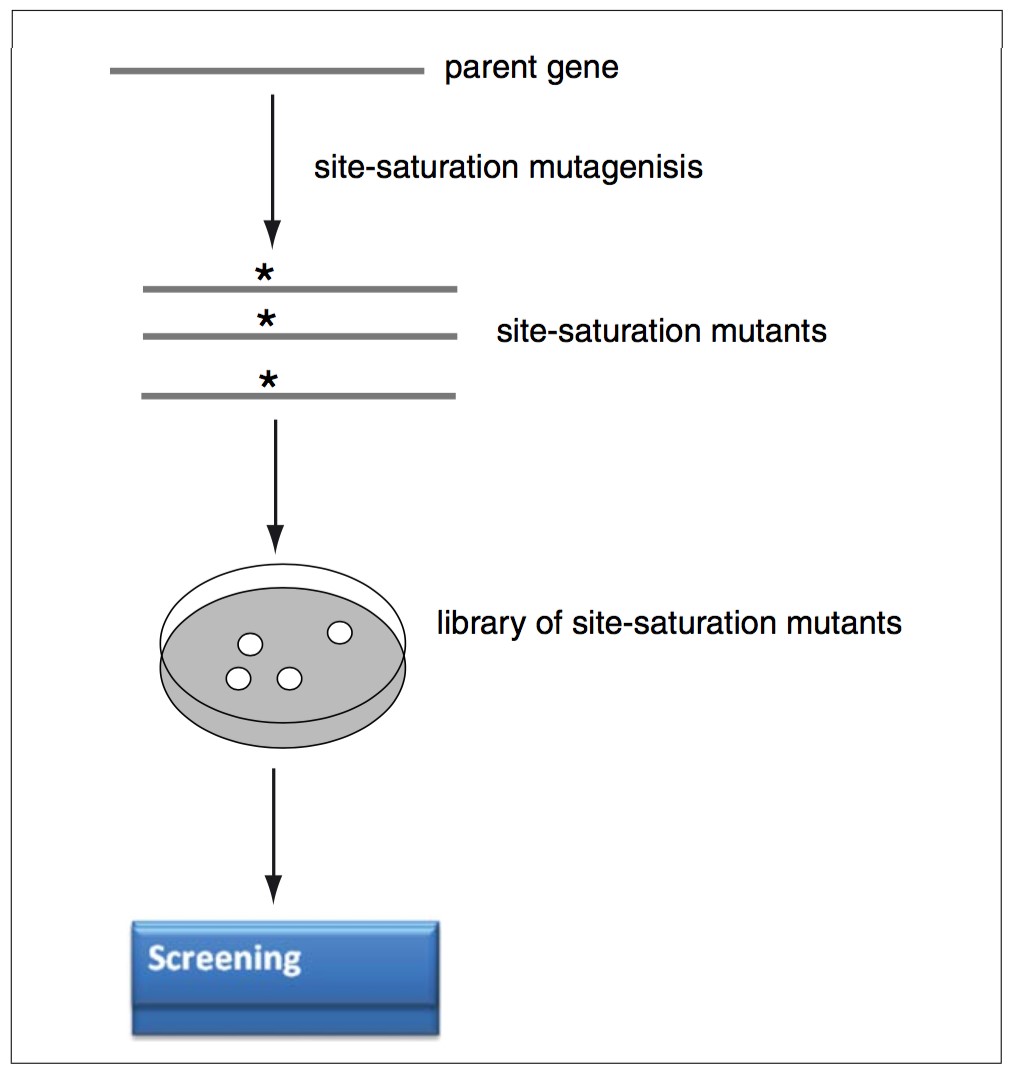 Figure 1. The main experimental steps of an in vitro–directed evolution process using site-saturation mutagenesis.
Figure 1. The main experimental steps of an in vitro–directed evolution process using site-saturation mutagenesis.
SSM can be carried out based on two oligonucleotide primers containing mutant condons with a mismatched sequence, complementary to the opposite strands of the double-stranded DNA template, are extended using polymerases. The wild type DNA plasmid template is then selectively degraded by DpnI endonuclease. Then, this synthetic plasmid can be directly transformed into competent E. coli strains and the nick in the DNA is repaired in vivo by the cell machinery to yield a mutated, circular plasmid. This procedure eliminates time-consuming subcloning, ligation, and single-stranded DNA rescue.
Creative Biostructure can provide several optionis for site-saturation mutagenesis services (such as sequential permutation) according to your needs. You can select to creative variants one or more codons, and receive constructs individually or pooled.
Our deliverables includes but not limited to:
- Variant constructs subcloned into the vector of your choice;
- Pooled constructs bulk-sequenced to verify mutation at the requested positions and the nonmutated regions intact;
- Delivery of all 19 non-WT variants at single or multiple site;
- Constructs sequenced for varification.
A wide range of applications we can provide by site-saturation mutegenesis:
- Change substrate specificity
- Identify beneficial or detrimental amino acid substitutions
- Identify active sites or receptor-binding sites
- Increase affinity, specificity, activity, heat stability, detergent tolerance
Please feel free to contact us to discuss your project!
Ordering Process
References:
E. G. Chronopoulou and N. E. Labrou (2011). Site-saturation Mutagenesis: A Powerful Tool for Structure-Based Design of Combinatorial Mutation Libraries. Current Protocols in Protein Science,UNIT 26.6. DOI: 10.1002/0471140864.ps2606s63
Saturated mutagenesis. (https://en.wikipedia.org/wiki/Saturated_mutagenesis)
