Membrane Protein Crystallization
In biological cells, membrane proteins (membrane proteins, MPs) are the most critical components to maintain cellular physiological processes and play an essential role in various cellular functions such as material transport, signal transduction, and intercellular recognition. MPs encoded in the human genome account for approximately 25% of the total proteome. These MPs are associated with various diseases, such as cystic fibrosis, atherosclerosis, Parkinson's disease, and Alzheimer's disease. Among them, transmembrane proteins are currently the most important drug targets, accounting for more than 60% of the known drug targets at this stage. The research and development of innovative drugs targeting transmembrane proteins has always been a research hotspot in the field of medicine.
Utilizing Lipid Cubic Phase for Membrane Protein Crystallization
Membrane protein molecules are amphiphilic, the hydrophobic part is close to the inside of the membrane, and the hydrophilic part is exposed outside the membrane. This lipophilic and hydrophilic structure makes it difficult to obtain highly ordered three-dimensional crystals as long as they leave the natural phospholipid bilayer hydrophobic environment, and their non-polar parts will form disordered polymers and pose a huge obstacle to the structural analysis of membrane proteins. The lipid cubic phase (LCP) has been widely used as a medium for crystallizing membrane proteins. Since the lipid cubic phase contains lipid bilayers, it can simulate biological membranes, which can help membrane proteins maintain their natural conformation. Lipid cubic phase technology has successfully aided the crystallization of multiple GPCRs and related drug development. Creative Biostructure's membrane protein research team has been actively developing and optimizing research methods for various membrane proteins, such as receptors, ion channels, and transporters. Crystallization of membrane proteins usually begins by testing a large number of possible crystallization reagents to determine initial conditions, which are then further optimized to obtain well-diffracting crystals.
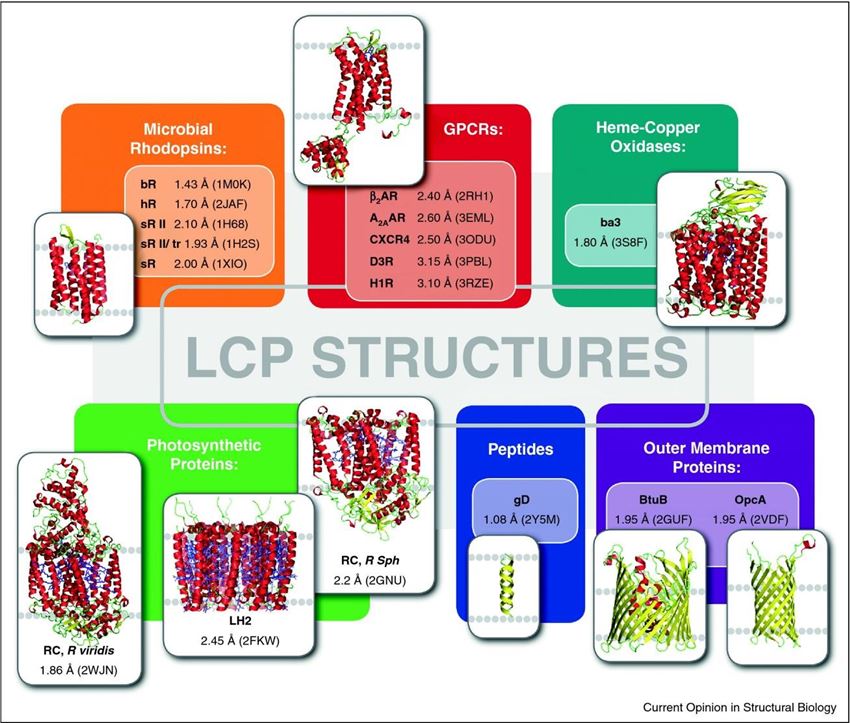 Figure 1. A gallery of protein and peptide structures obtained by LCP crystallization.
Figure 1. A gallery of protein and peptide structures obtained by LCP crystallization.
Our Service Process for Membrane Protein Crystallization
| procedure | Information |
| Step 1 | Sequence optimization, gene synthesis |
| Step 2 | Protein expression and purification |
| Step 3 | High-throughput screening of protein crystal conditions |
| Step 4 | Protein synchrotron radiation diffraction data collection |
| Step 5 | Protein-compound co-crystallization & Fragment screening |
| Step 6 | Protein crystal data analysis |
Advantages of Our Membrane Protein Crystallization Service
Creative Biostructure has rich experience in membrane protein research, and the advantages of our service include the following:
- We optimized the method based on sitting drop vapor phase diffusion lipid cubic phase crystallization and X-ray micro focusing and established an effective path for analyzing the structure of new membrane proteins.
- We have extensive experience in resolving novel protein structures from scratch.
- Our platform enables membrane proteins to be crystallized while maintaining their native conformation and retaining their function and activity.
- We can explore other biochemical, chemical, and physical parameters in crystallization screening experiments.
Creative Biostructure's efficient protocols and extensive process optimization significantly increase the success rate, and we work closely with our customers to provide tailored strategies for the structure determination of membrane proteins of interest. Please feel free to contact us for a detailed quote.
Ordering Process
Reference
- Cherezov V. Lipidic cubic phase technologies for membrane protein structural studies. Current Opinion in Structural Biology. 2011, 21(4): 559-566.
