Real-time Protein NMR Service
The 3D structure, dynamics, and interactions of proteins are important elements in chemical and structural biology research. How to develop high-resolution methods to elucidate the mechanism of action and functional relationships of macromolecules in an intracellular environment is an important problem that needs to be solved so far.
NMR techniques have the unique advantage of being able to resolve not only the 3D structure of protein molecules but also the kinetic properties of ligand-receptor interactions and protein conformational changes under near-physiological conditions. It also provides information on the fine structure of macromolecules at the nucleus and electron levels to understand the mechanism of macromolecular motility in the intracellular environment.
How We Offer Our Services
Creative Biostructure relies on the structural biology tool, NMR to characterize time-dependent protein interactions in living cells at atomic resolution through real-time protein NMR services.
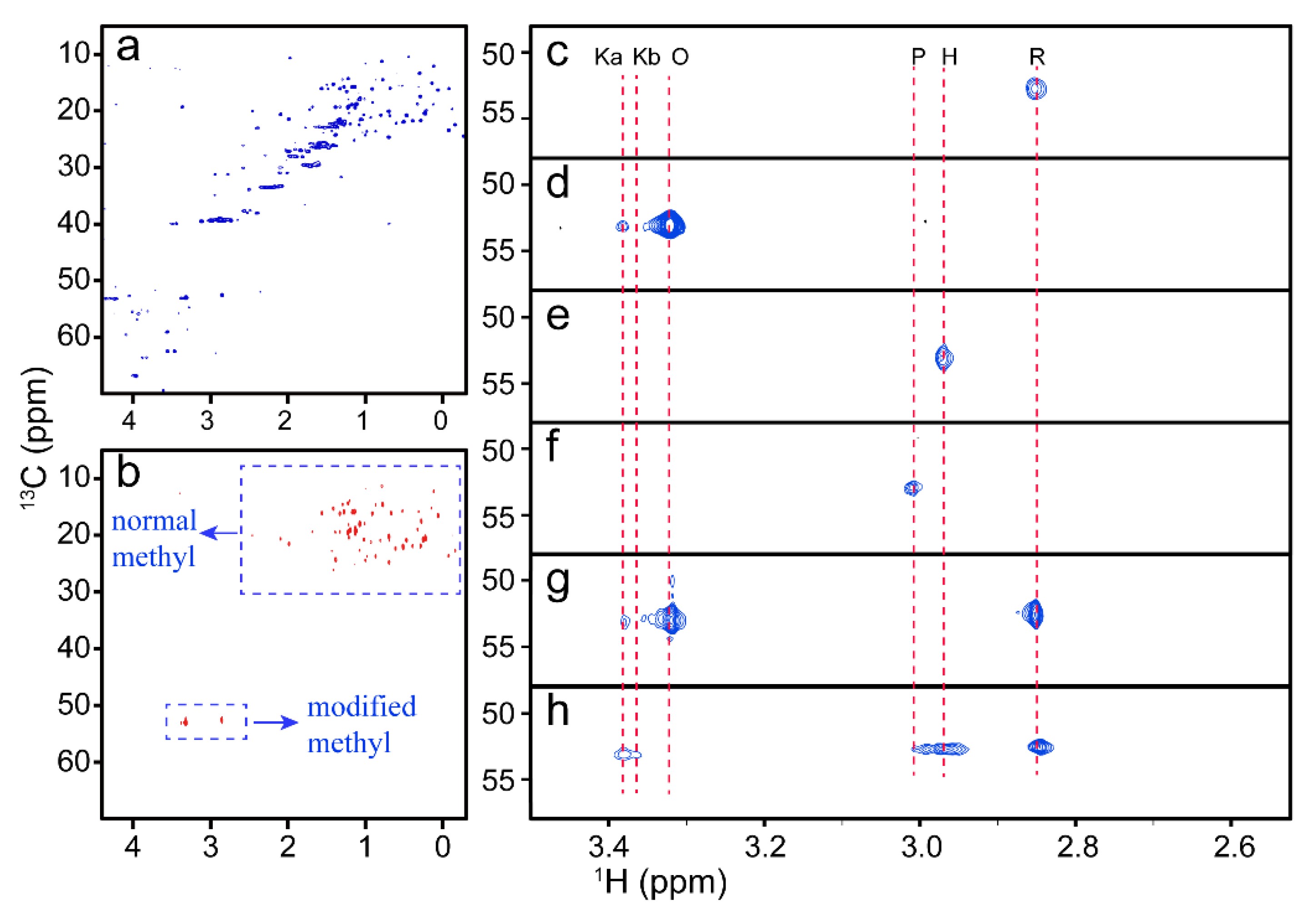 Figure 1. NMR spectra of cyt c with different conformations. (Zhan, J.; et al. 2021)
Figure 1. NMR spectra of cyt c with different conformations. (Zhan, J.; et al. 2021)
- One-dimensional H-spectroscopy together with two-dimensional homonuclear H-spectroscopy can be used to understand the overall protein folding, whether the sample is aggregated, etc. without protein labeling.
- Two-dimensional heteronuclear experiments can be used to study protein-ligand interactions. For proteins with existing backbone attribution, the site of action of the protein molecule can be measured directly. For protein molecules without backbone attribution, it is possible to understand whether the protein molecule interacts with the ligand or not.
- Triple resonance 3D NMR is mainly used for main side-chain chemical shift attribution, and 3D NOE experiments are mainly used to obtain spatial structure information, which can be combined with main side-chain chemical shift attribution to perform 3D structure calculation.
Sample Requirements
The molecular weight of the protein molecule used for detection needs to be less than 30kDa, the concentration should be greater than 0.5mM, and the volume should be 450uL-500uL. The specificity is also related to the specific structure of the protein molecule itself and the information customers need to obtain.
- For 1D NMR, the molecular weight can be extended to 100kDa and the concentration can be as low as 0.01mM.
- For 2D NMR, the molecular weight can be up to 50kDa and the concentration can be as low as 0.1mM.
- For 3D NMR, the molecular weight must be less than 30kDa, preferably less than 25kDa, and the concentration must be at least 0.5mM.
The 2D and 3D analysis needs to be done in isotopically labeled samples. In addition, it is recommended that you perform a secondary structure prediction of the target protein before considering using NMR to study the protein. This is because it may affect the final signal intensity and spectral resolution.
Our Advantages
- Information about the structural and dynamic characteristics of proteins with short half-lives can be resolved.
- It is possible to obtain atomic-resolution protein structure, dynamics, and interaction information in living cells.
- Our service enables complete and accurate attribution of NMR peak information of abundant protein molecules.
As a leader in the field of NMR for the resolution of biological macromolecules, it is our goal to help our customers solve problems in the application of NMR to protein research. Please feel free to contact us to start a new project collaboration.
Ordering Process
Reference
- Zhan J.; et al. NMR reveals the conformational changes of cytochrome C upon interaction with cardiolipin. Life. 2021, 11(10): 1031.
