Epitope Mapping by 3D-EM
Epitope mapping is the experimental identification of the binding sites of antibodies on their target antigens. The mapping of epitopes is a fundamental step in the development of novel therapeutics, vaccines, and diagnostics. Epitopes are generally divided into two main types: linear and conformational. Linear epitopes are formed by a continuous sequence of amino acids in a protein, while conformational epitopes are composed of amino acids that are discontinuous in the protein sequence but are brought together upon three-dimensional protein folding. Many therapeutic monoclonal antibodies recognize conformational epitopes of target antigens, which are often represented by membrane proteins, receptors, or multi-subunit proteins.
High-resolution structure of the antibody/antigen (Ab/Ag) complex is the key to accurate epitope mapping. Historically, X-ray crystallography has been used to provide high-resolution information of antibody-antigen interactions, but it is costly and time-consuming, and requires large amounts of purified samples. To avoid these problem, Creative Biostructure is devoted to developing electron microscopy (EM) as a new method of mapping epitopes by single-particle analysis and three-dimensional reconstruction (3D-EM). Taking advantage of the EM platform, our scientists perform epitope mapping of antibody-antigen interactions with the following workflow:
- Purified antigens are incubated with high-affinity Fabs and the complex purified by SEC.
- The resulting complexes are then deposited on EM grids and subjected to either negative staining or cryo-EM preparation.
- The prepared grids are then imaged on a Transmission Electron Microscope (TEM).
- Following data collection, the resulting images are used to produce a dataset of 104-105 particles.
- This dataset is then processed to obtain reference-free 2D classes and 3D reconstructions of the antigen-antibody complexes.
- Lastly, the crystal structures or homology models of Fabs and antigens are computationally docked into the 3D reconstructions, allowing for the identification of the residues at the binding interface.
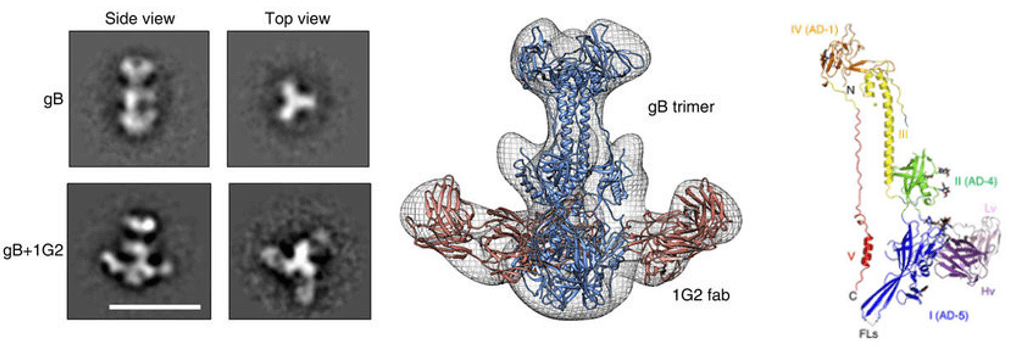 Figure 1. Epitope mapping by 3D-EM
Figure 1. Epitope mapping by 3D-EM
The advantages of using our EM platform for epitope mapping include sample-saving, rapid turn-around time, and cost-effective. Moreover, the entire process can be run manually, or in an automated way that can be completed within a few hours on a high-performance computer cluster. Creative Biostructure promises to work closely with our customers to provide tailored strategies for the epitope mapping of antibody-antigen complexes using 3D-EM technique.
Besides NMR, Creative Biostructure offers a field of structural biology for mapping protein epitopes, such as Epitope Mapping by crystallography and 3D-EM. Applications include epitope mapping, antibody engineering (affinity maturation and humanization), and protein engineering.
Please feel free to contact us for a detailed quote.
Ordering Process
References:
- Chandramouli S, et al. (2015) “Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody”. Nat Commun 6:8176.
- Lee H, et al. (2015) “A cryo-electron microscopy study identifies the complete H16.V5 epitope and reveals global conformational changes initiated by binding of the neutralizing antibody fragment”. J Virol 89(2):1428-1438.
