Isotope Labeled Protein Production
Creative Biostructure is pleased to offer a custom stable isotope labeling service for proteins with expression and purification, as well as for nucleic acids. We specialize in labeling proteins selectively and uniformly with stable isotopes, typically 1H, 2H, 15N, and 13C. Also, we can label proteins based on specific requirements. All Creative Biostructure products undergo a strict quality control process, including nuclear magnetic resonance (NMR) analysis.
Isotopic labeling has played a crucial role in the development of solid-state NMR spectroscopy that determines the structures of proteins that reside in membranes, aggregates, or other types of assemblies. The use of advanced isotopic labeling methods enables near 100% incorporation of 1H, 2H, 15N, and 13C spins in strategic positions of target samples with limited scrambling. It simplifies NMR spectra and helps investigate interactions, structures, and dynamics of large and complex biological systems. These sophisticated labeling schemes often allow a significant enhancement in the signal-to-noise of NMR experiments and, concomitantly, a speed-up of their acquisition.
Generally, there are several approaches to express proteins and peptides: cell-based expression system, cell-free expression system, and chemical synthesis. The most widely used cell-based expression systems are bacteria, yeasts, and insect cells (Table 1).
Table 1. Cell-based isotope labeled protein production system.
| E. coli | Yeast | Insect cells | Mammalian cells | |
| Ubiquitous labeling | 15N, 13C, 2H | 15N, 13C | 15N, 13C- labeling is possible | 15N, 13C- labeling is possible |
| Medium | Ammonium salts as 15N-source, glucose as 13C-source | Ammonium salts as 15N-source, methanol as 13C-source | Minimal media | - |
Below are three main isotope labeling approaches that Creative Biostructure provides:
- Specific isotopic labeling: Placement of a single label (2H, 13C, or 15N) in a specified location in a polypeptide. Specific labeling is accomplished by incorporating a labeled amino acid through chemical synthesis, which restricts this approach to relatively small polypeptides.
- Selective isotopic labeling: Biosynthetic incorporation of a single type of labeled amino acid with all of the others unlabeled. Selective labeling is widely used to assist resonance assignments in both solution and solid-state NMR of aligned samples. It serves as a method to simultaneously assign resonances and determine structure.
- Uniform isotopic labeling: Biosynthetic labeling of all carbon, nitrogen, or hydrogen sites with stable isotopes. Uniform labeling of proteins with 15N is particularly convenient because of the strategic locations of nitrogens in the backbone and the absence of homonuclear couplings due to the intervening carbons.
Creative Biostructure can also perform combinations of specific and selective labeling, as well as combinations of uniform and selective labeling. The former is crucial for the solid-state NMR that measures specific distances between strategically placed pairs of nuclei in unoriented protein samples. The latter enables the resonances from one type of amino acids to compare spectra from uniformly and selectively labeled protein samples in both solution and solid-state NMR studies of proteins. Besides, segmental labeling, fractional labeling, deuteration and paramagnetic labeling are supported by us.
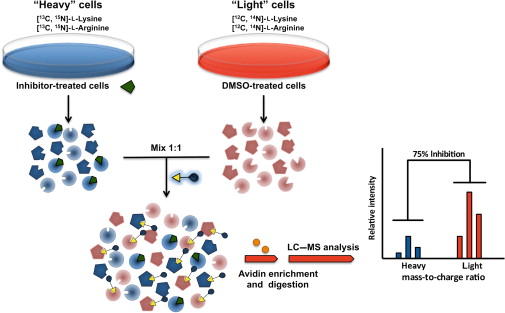 Figure 1. Stable isotope labeling by
amino acids in cell culture (SILAC) enables in vivo incorporation of a label into proteins for mass spectrometry-based quantitative proteomic
analysis (Ong et al., 2002).
Figure 1. Stable isotope labeling by
amino acids in cell culture (SILAC) enables in vivo incorporation of a label into proteins for mass spectrometry-based quantitative proteomic
analysis (Ong et al., 2002).
The isotope labeling platform of Creative Biostructure is devoted to the large-scale production of proteins uniformly or specifically enriched in stable isotopes (1H, 2H, 15N, and 13C) for biomolecular NMR spectroscopy studies. The optimized protocols and adequate isotopically labeled materials are developed at Creative Biostructure. Please feel free to contact us for a detailed quote.
Ordering Process
References
- Boisbouvier J, Kay L E. Advanced isotopic labeling for the NMR investigation of challenging proteins and nucleic acids. Journal of Biomolecular NMR. 2018: 115-117.
- Ong S E, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & Cellular Proteomics. 2002, 1(5): 376-386.
