Exosomal Circular RNA (circRNA) Sequencing Service
Exosomal circular RNAs (circRNAs), as novel disease biomarkers and regulatory molecules, offer advantages such as high stability and strong specificity. Creative Biostructure provides comprehensive exosomal sequencing analysis services, accurately deciphering the expression profiles and functions of circRNAs within exosomes to facilitate disease biomarker discovery and mechanistic research.
What are Exosomal circular RNAs and Why Sequence Them?
Circular RNAs (circRNAs) are a class of non-coding RNAs characterized by a covalently closed loop structure, endowing them with high stability and strong tissue specificity. Exosomes are nano-sized vesicles secreted by cells, carrying a variety of biomolecules, including circRNAs. Exosomal circRNAs hold significant potential as biomarkers for disease diagnosis and therapeutic targets, garnering considerable attention in research areas such as cancer and neurodegenerative diseases.
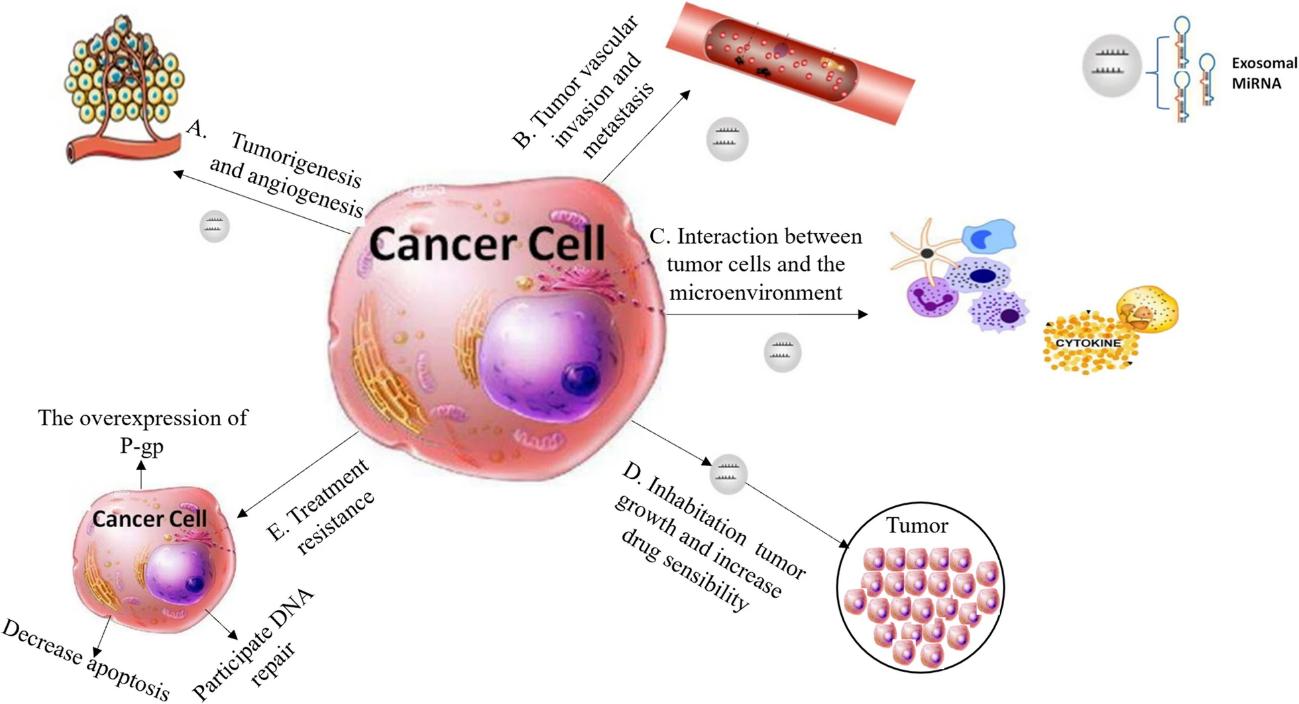
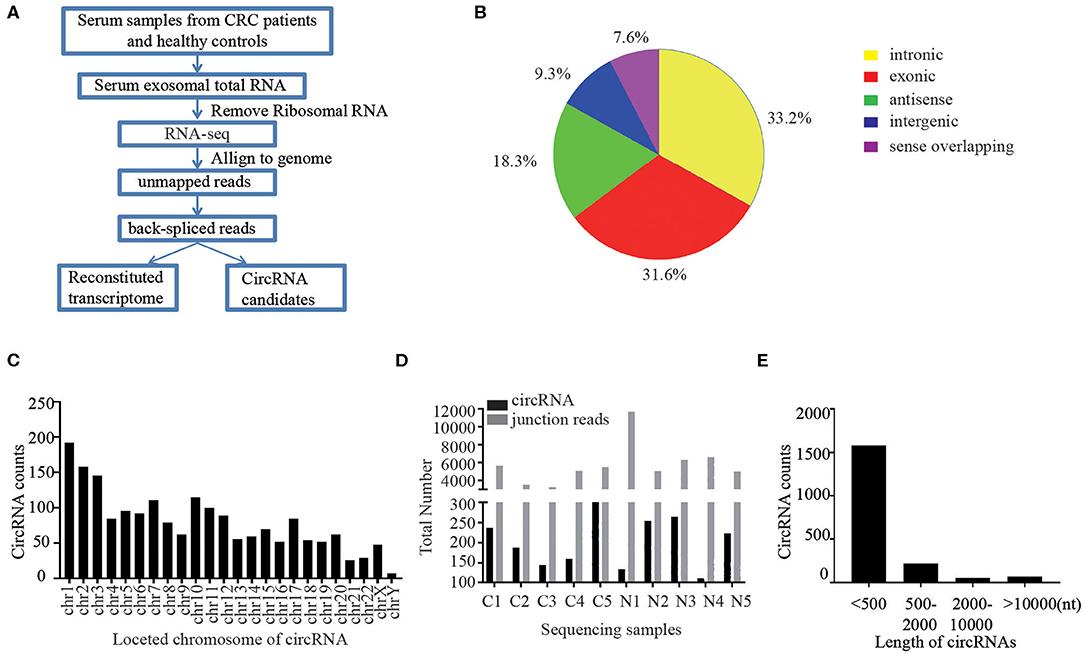 Figure 1. Profiling of circular RNAs in human serum exosomes. (Xie, Y. et al., 2020)
Figure 1. Profiling of circular RNAs in human serum exosomes. (Xie, Y. et al., 2020)
Our Exosomal circRNA Sequencing Services
Our exosome circular RNA sequencing service specializes in precisely capturing regulatory circular RNA molecules within exosomes. Our expert team provides comprehensive experimental services from sample quality control to library construction, complemented by multidimensional bioinformatics analysis to deeply uncover biological significance. From rigorous quality control to personalized result interpretation, we deliver high-quality data support throughout the process for scientific needs such as disease biomarker discovery and mechanism research.
| Services | Descriotion |
|---|---|
| Exosome Isolation and Exosome Characterization | Exosomes are isolated using various methods such as ultracentrifugation and magnetic bead-based separation, and are characterized by NTA, electron microscopy, and Western blot.
|
| circRNA Sequencing | Employing high-throughput sequencing technology to comprehensively analyze the expression profiles of circRNAs within exosomes and identify novel circRNA molecules.
|
| Bioinformatics Analysis | We provide comprehensive circRNA data analysis services, including identification, quantification, differential analysis, and functional prediction.
|
Service Workflow of Exosomal circRNA Sequencing Analysis
We provide a standardized service workflow to ensure efficient and high-quality completion of projects, offering robust support for your research.
Sample Preparation and Exosome Isolation
Receive and process various biological samples, such as cell supernatants, plasma, serum, etc. Isolate and purify intact exosomes from samples using standard methods including ultracentrifugation or size-exclusion chromatography.
RNA Extraction and Quality Control
Total RNA is extracted from isolated exosomes using an optimized kit to maximize preservation of circular RNA. Additionally, we perform rigorous RNA quality control to ensure sample integrity meets library preparation standards.
Circular RNA Library Construction and Sequencing
Ribosomal RNA is depleted using specialized kits to remove abundant rRNA species, thereby enriching non-coding RNAs including circRNAs. A strand-specific library is then constructed, a step that is critical for the subsequent accurate identification of back-splicing junctions in circRNAs. Finally, high-throughput sequencing is performed on platforms such as the Illumina NovaSeq with a PE150 configuration, generating raw data of sufficient depth for comprehensive analysis.
Bioinformatics Analysis
We offer a variety of bioinformatics analysis services, including but not limited to the following:
| Data Quality Control and Alignment | This step uses FastQC for raw data quality control, followed by alignment of high-quality sequencing reads to the reference genome using software such as STAR. |
| circRNA Identification and Quantification | Specialized algorithms including CIRI2 and find_circ are applied to accurately identify circRNAs featuring back-splicing junctions, followed by expression quantification. |
| Differential Expression Analysis | Statistical tools like DESeq2 are utilized to screen for differentially expressed circRNAs between case and control groups. |
| Functional Enrichment Analysis | GO and KEGG pathway enrichment analyses are performed on host genes or predicted target genes of differentially expressed circRNAs to reveal their potential biological functions. |
| ceRNA Network Construction | Based on the competitive endogenous RNA theory, a regulatory network consisting of circRNAs, miRNAs, and mRNAs is constructed to further elucidate potential molecular mechanisms. |
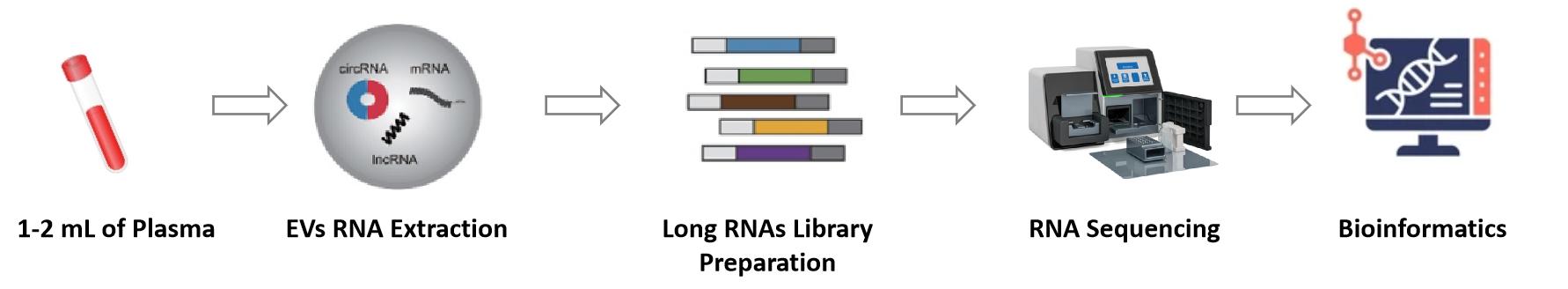
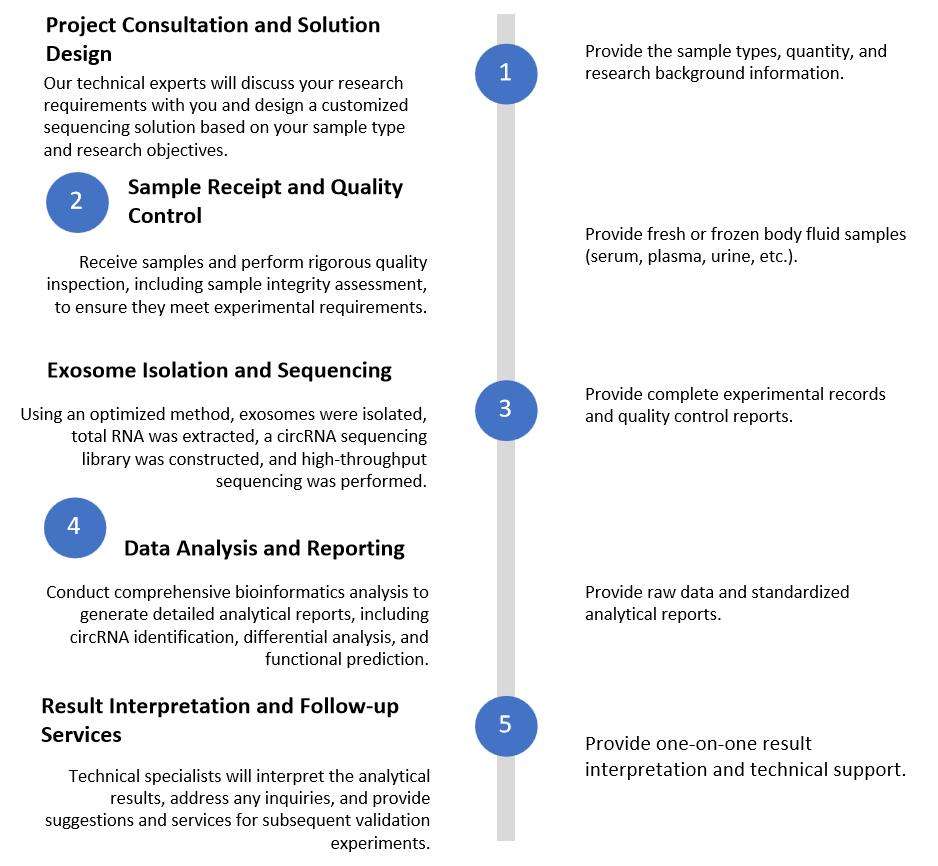 Figure 2. Exosomal circRNA Sequencing Services Flow. (Creative Biostructure)
Figure 2. Exosomal circRNA Sequencing Services Flow. (Creative Biostructure)
Advantages of Our Exosomal circRNA Sequencing Services
We possess leading sequencing platforms and a professional bioinformatics analysis team to provide you with high-quality exosomal circular RNA sequencing services.
- High-Sensitivity Detection
Utilizing advanced sequencing technology and optimized experimental workflows, this method detects low-abundance exosomal circRNAs, ensuring comprehensive capture of expression profile information.
- Professional Exosome Isolation
By leveraging the advantages of multiple isolation methods, we obtain exosomes with high purity and integrity, thereby minimizing contamination from non-exosomal RNA.
- Comprehensive Data Analysis
We offer a comprehensive analytical pipeline ranging from basic analysis to advanced functional prediction, encompassing circRNA identification, quantification, differential analysis, and ceRNA network construction.
- Rigorous Quality Control
Rigorous quality control is implemented at each stage, from sample reception through experimental procedures to data analysis, to ensure the reliability and reproducibility of the results.
Sample Scope & Submission Guide
| Sample Type | Minimum Input Amount | Applicable Scenarios |
|---|---|---|
| Serum/Plasma | ≥200 μL | Disease Biomarker Research |
| Cell Culture Supernatant | ≥5 mL | Intercellular Communication Mechanisms |
| Urine | ≥10 mL | Diagnostic Research |
| Tissue Homogenate Supernatant | ≥100 mg | Tumor Microenvironment Research |
Final Deliverables
- Raw sequencing data (FASTQ files, compatible with GEO/SRA submission)
- Quality control report (RNA integrity, sequencing depth, duplication rate)
- circRNA dataset (ID, chromosome location, back-splicing site, expression level)
- Differential circRNA list (with log2FC, p-value, FDR)
- Visualization files (volcano plots, heatmaps, circRNA structure diagrams)
- Comprehensive analysis report (methods, results, preliminary interpretation)
Case Study
Case: Circular RNA Sequencing Reveals Serum Exosome Circular RNA Panel for High-Grade Astrocytoma Diagnosis
Background
Circular RNAs (circRNAs) have potential as tumor biomarkers due to their stability, conservation, and tissue specificity. However, the landscape and characteristics of exosomal circRNAs in high-grade astrocytomas (HGAs) remain to be elucidated. This study reveals a comprehensive circRNA profile in both exosomes and cells of HGAs.
Methods
- CircRNA deep sequencing and bioinformatics approaches were used to generate a circRNA profiling database and analyze the features of HGA cell circRNAs and HGA cell-derived exosome circRNAs.
- Exosome circRNA expression in the serum and tissues of healthy individuals and patients with HGA was detected using reverse transcription-quantitative PCR.
Conclusion
This study identified circRNA profiles, the features of circRNAs in primary HGA cells and cell-derived exosomes, and confirmed a serum exosome circRNA panel (hsa_circ_0003828, hsa_circ_0075828, and hsa_circ_0002976) for liquid biopsy in early HGA screening.
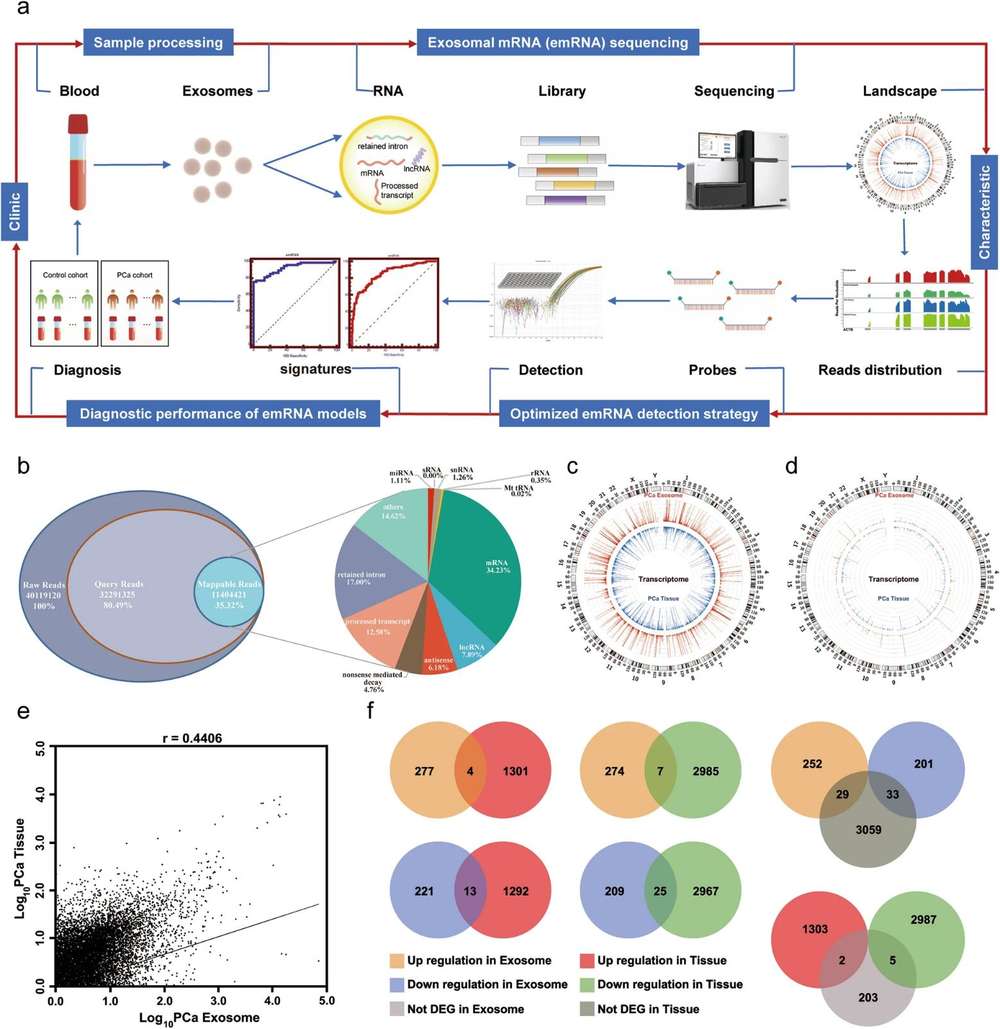
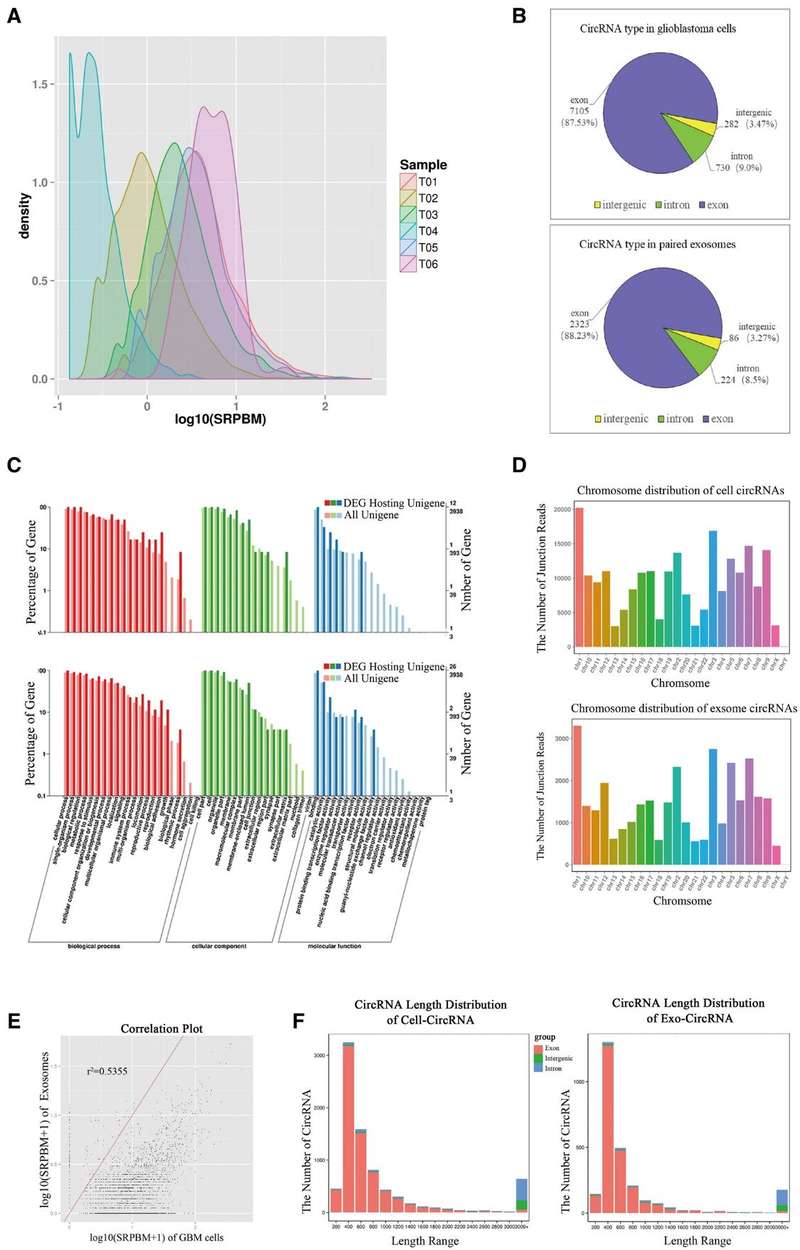 Figure 3. Profiling characteristics of primary HGA-derived exosome circRNAs. (Li, P. et al., 2022)
Figure 3. Profiling characteristics of primary HGA-derived exosome circRNAs. (Li, P. et al., 2022)
Creative Biostructure is committed to providing clients with professional exosomal circRNA sequencing services. We offer comprehensive support throughout the entire process, from precise experimental procedures to in-depth bioinformatics analysis, ensuring you obtain clear, reliable, and publication-ready high-quality data. Contact us now for more details.
References
- Xie Y, Li J, Li P, et al. RNA-Seq profiling of serum exosomal circular RNAs reveals Circ-PNN as a potential biomarker for human colorectal cancer. Frontiers in oncology. 2020, 10: 982.
- Li, P, Xu Z, Liu T, et al. Circular RNA sequencing reveals serum exosome circular RNA panel for high-grade astrocytoma diagnosis. Clinical chemistry. 2022, 68(2): 332-343.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.