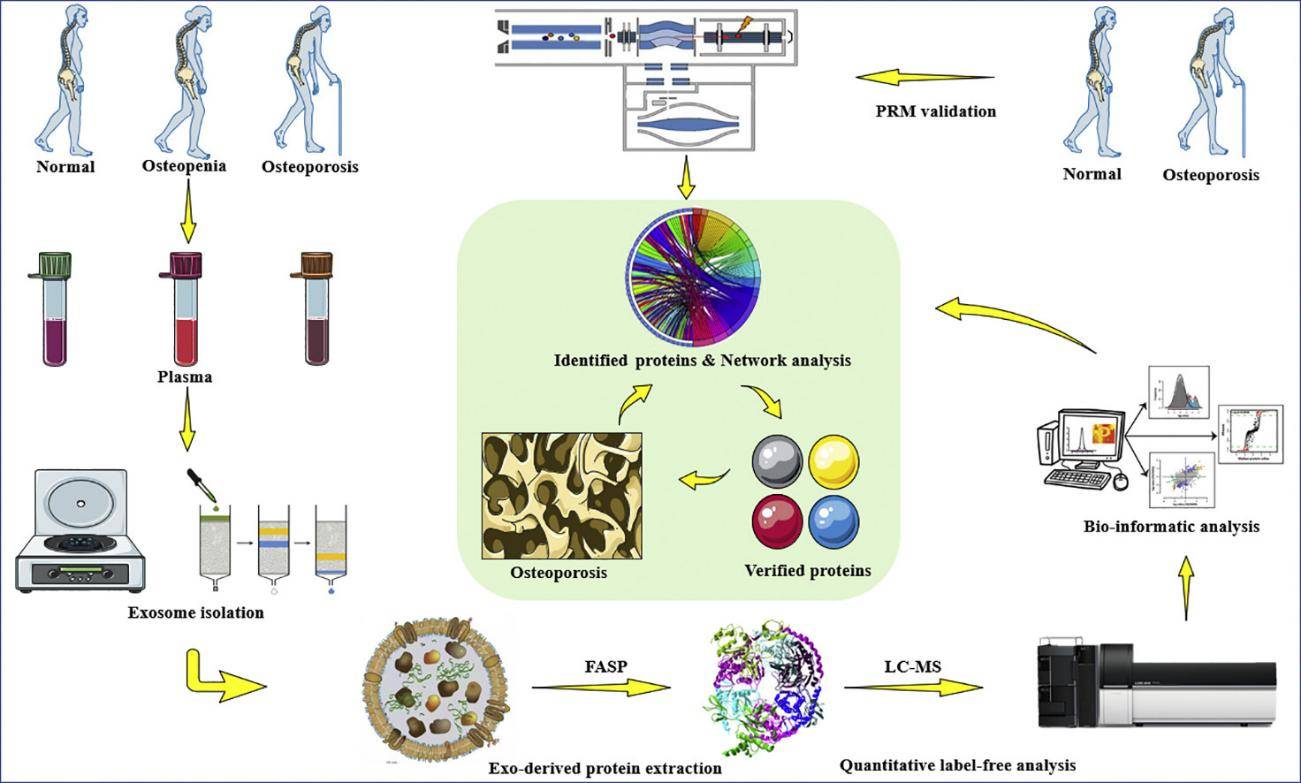
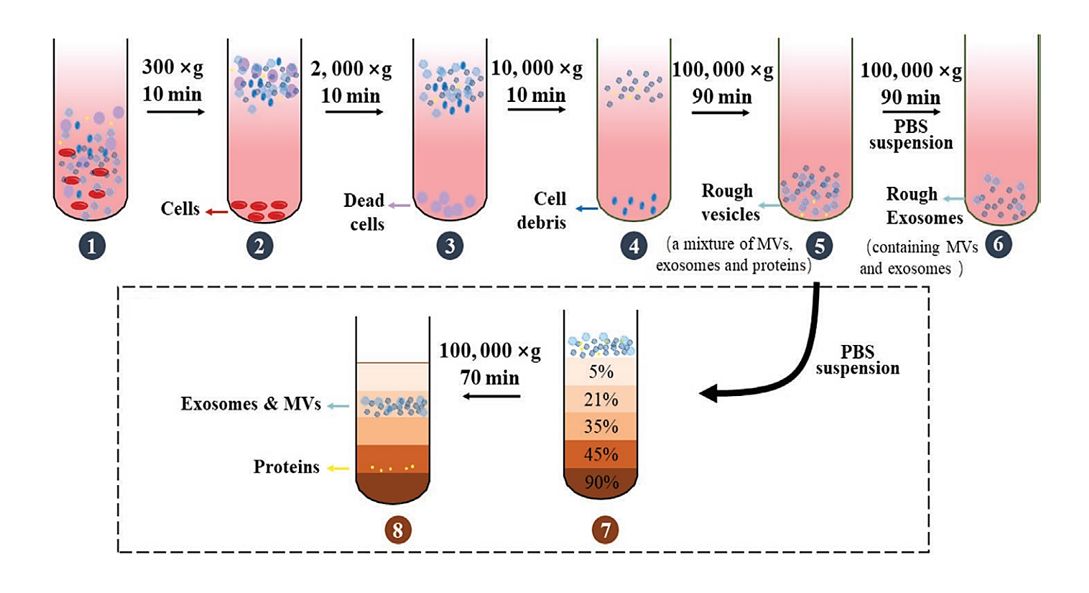
Exosome Protein Profiling Service with Olink Proteomics Technology
Due to the limited sample volume of EVs, detecting protein content requires technologies with high sensitivity and accuracy. Olink's PEA technology has gained popularity in EV research for its minimal sample consumption and exceptional detection sensitivity. Creative Biostructure's Olink proteomics service effectively addresses the limitations of conventional exosomal protein mass spectrometry by integrating high-purity exosome isolation with the proprietary Proximity Extension Assay (PEA) technology. Our combined approach enables ultrasensitive and highly specific detection of hundreds of exosomal proteins under extremely low sample input conditions, paving a new path for exosome research and clinical translation.
Introduction to Olink Technology and Its Application Value in Exosome Research
Exosomes carry specific proteins, nucleic acids, and other information from their source cells, making them a valuable source of biomarkers for disease diagnosis. However, due to their small size and limited contents, detecting low-abundance proteins has remained a technical challenge. The Olink PEA technology demonstrates significant advantages in this regard:
- Unveiling the Exosomal Proteome
Olink's high sensitivity and multiplex detection capability allow researchers to deeply explore the protein composition of exosomes from limited clinical samples (such as small volumes of serum or plasma), enabling the discovery of characteristic protein expression profiles associated with diseases.
- Tracing Cell Origins and Functions
By analyzing specific membrane proteins on the surface of exosomes (such as CD63, CD9, etc.) and their cargo proteins, Olink technology helps infer the cellular origin of exosomes and reveals their functions in processes like intercellular communication and immune regulation.
- Discovery and Validation of Disease Biomarkers
The high sensitivity and stability of Olink technology make it well-suited for biomarker discovery and validation. Particularly in the field of inflammation research, Olink offers specialized panels that can efficiently capture a wide range of low-abundance cytokines. These cytokines play critical roles in immune and inflammatory responses but are difficult to effectively detect using traditional mass spectrometry methods.
Exosome Protein Profiling with Olink Proteomics Technology
Olink Proteomics technology, particularly its unique Proximity Extension Assay (PEA), provides a powerful tool for proteomic research of minimal samples such as exosomes. Our technology enables ultra-high sensitivity, high specificity, and highly multiplex protein detection and quantification, helping clients gain deeper insights into disease mechanisms, discover biomarkers, and advance the development of precision medicine.
Our exosomal protein analysis service, based on Olink Proteomics Technology, features the following characteristics:
| Features | Description | Advantages |
|---|---|---|
| Ultra-high sensitivity | The detection sensitivity can reach the fg/mL (femtogram per milliliter) level. | Capable of effectively detecting low-abundance proteins (such as cytokines) in exosomes that are difficult to detect using traditional methods. |
| Minimal sample requirements | Only 1-6 μL of sample volume is required for testing. | Ideal for research involving precious or trace samples such as exosome lysates, cerebrospinal fluid, serum, and similar specimens. |
| High multiplexing capability | Simultaneously detects 48-3072 protein biomarkers. | Enable high-throughput screening to obtain extensive protein information in a single experiment, revealing complex protein regulatory networks. |
| Wide dynamic range | Dynamic range spanning 10 orders of magnitude. | Capable of simultaneously and accurately quantifying proteins at high, medium, and low abundances within samples, preventing signal saturation or loss. |
| High specificity and accuracy | By employing a dual-antibody pairing to recognize target proteins and integrating DNA barcoding technology, this approach effectively avoids antibody cross-reactivity and signal crosstalk issues commonly encountered in multiplex immunoassays. | Ensure the reliability and accuracy of test results while reducing the false positive rate. |
| Reproducibility | High technical redundancy. | Ensure data stability and reproducibility. |
| Compatible with multiple sample types | Suitable for various sample types including serum, plasma, cerebrospinal fluid, urine, cell culture supernatant, and exosome lysate. | Provides a unified, reliable proteomics analysis platform for samples from diverse sources, with particular expertise in detecting low-abundance proteins in bodily fluids. |
How Exosome Protein Profiling Works with Olink Proteomics Technology?
Olink's PEA technology elegantly combines the specificity of immunoassays with the sensitivity of nucleic acid amplification. Its core workflow is illustrated in the figure below:
Antibody-Oligonucleotide Probes
Olink pairs of antibodies, each labeled with a unique oligonucleotide probe, bind specifically to target proteins on the exosome.
Proximity Event
When both antibody probes bind to the same target protein, the DNA oligonucleotides are brought into close proximity.
DNA Amplification
A connector DNA Oligonucleotide allows the linked probes to form a detectable DNA template that can be amplified via real-time PCR or NGS.
Signal Conversion
The amplified DNA signal is directly proportional to the amount of the target protein.
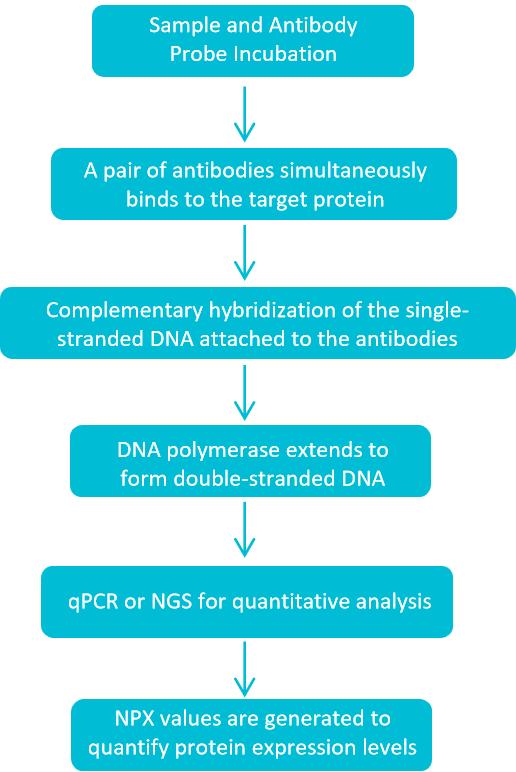 Figure 1. Exosome Protein Profiling Workflow with Olink Proteomics Technology. (Creative Biostructure)
Figure 1. Exosome Protein Profiling Workflow with Olink Proteomics Technology. (Creative Biostructure)
Data Analysis & Interpretation
We transform raw Olink data into actionable insights through multidimensional analysis:
| Analysis Items | Analysis Content |
|---|---|
| Basic Analysis | Protein concentration matrix, inter-sample CV calculation (<10% for technical replicates), missing value imputation. |
| Differential Analysis | Volcano plots, heatmaps, and fold-change (FC) analysis to identify disease-associated exosome proteins. |
| Biological Interpretation | GO/KEGG pathway enrichment (e.g., "exosome-mediated signaling," "PI3K-AKT pathway"), protein-protein interaction (PPI) networks. |
| Clinical Translation | ROC curve analysis (AUC calculation) for biomarker validation, correlation with clinical outcomes (e.g., survival, treatment response). |
Deliverables
- QC report (exosome purity/morphology data, Olink assay quality metrics).
- Raw data (Excel/FASTQ files) and processed data matrix.
- Comprehensive analysis report with visualizations and biological interpretations.
- Methodology document (for manuscript submission, compliant with Olink's technical standards).
Case Study
Case: Characterization of extracellular vesicles in metastatic urothelial cancer patients to discover protein signatures linked to therapeutic outcomes
Background
Patients with metastatic urothelial carcinoma (mUC) typically have a poor prognosis, making early prediction of systemic treatment response critical for improving patient outcomes. This study investigated the proteome of extracellular vesicles (EVs) isolated from consecutive plasma samples in a cohort of mUC patients receiving combination therapy with vinorelbine and sorafenib in a Phase I trial. The isolated EVs were of exosomal size and expressed exosomal markers CD9, TSG101, and SYND-1. The study found no association between the number of EVs per milliliter of plasma at baseline and progression-free survival (PFS). Proteomic analysis of the EVs using antibody-based adjacent extension analysis with the Olink Oncology II panel revealed heterogeneous protein expression patterns.
Key Findings
- A protein signature composed of SYND-1, TNFSF13, FGF-BP1, TFPI-2, GZMH, ABL1, and ERBB3 is hypothesized to be associated with progression-free survival (PFS) in metastatic urothelial carcinoma (mUC).
- Protein features associated with optimal treatment response were identified, including FR-alpha, TLR3, TRAIL, and FASLG.
- Machine learning classification algorithms also identified several markers associated with PFS or optimal treatment response characteristics.
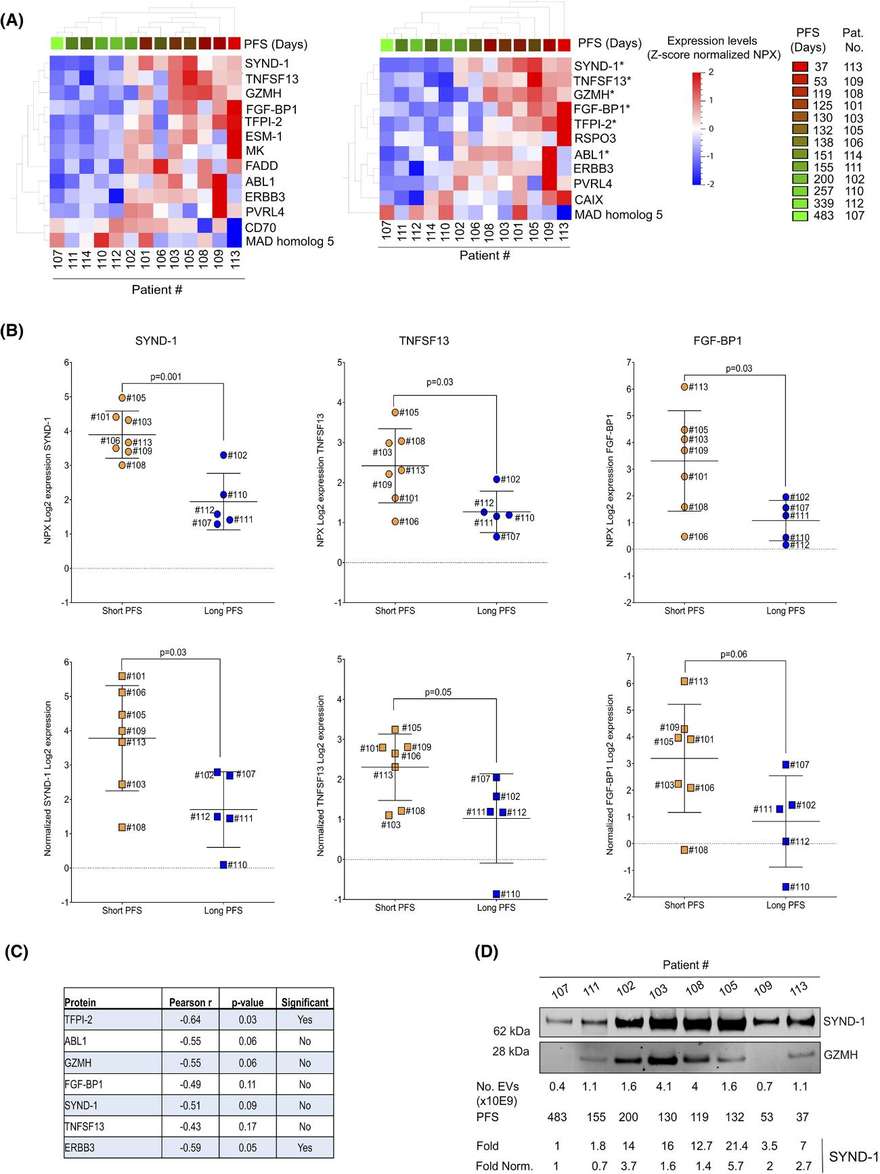 Figure 2. The relationship between extracellular vesicle protein analysis and progression-free survival (PFS). (Viktorsson K, et al., 2022)
Figure 2. The relationship between extracellular vesicle protein analysis and progression-free survival (PFS). (Viktorsson K, et al., 2022)
Creative Biostructure is dedicated to providing clients with professional exosome protein analysis services. Our team of Olink-certified specialists and exosome researchers will provide tailored solutions to accelerate your research. Please contact us for a customized experimental design plan. We will be happy to assist you.
Reference
- Viktorsson K, Hååg P, Shah C H, et al. Profiling of extracellular vesicles of metastatic urothelial cancer patients to discover protein signatures related to treatment outcome. Molecular oncology. 2022, 16(20): 3620-3641.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.