Exosome Omics Analysis Services for Mechanism-Driven Discovery
Not sure which omics strategy best answers your exosome question? Creative Biostructure provides end-to-end exosome multi-omics analysis services—from proteomics and lipidomics to miRNA/mRNA profiling and integrative bioinformatics—built around MISEV2023-aligned workflows.
Exosome Solutions for Your Proceeding Research
Download Brochure
What Is Exosome Omics Analysis?
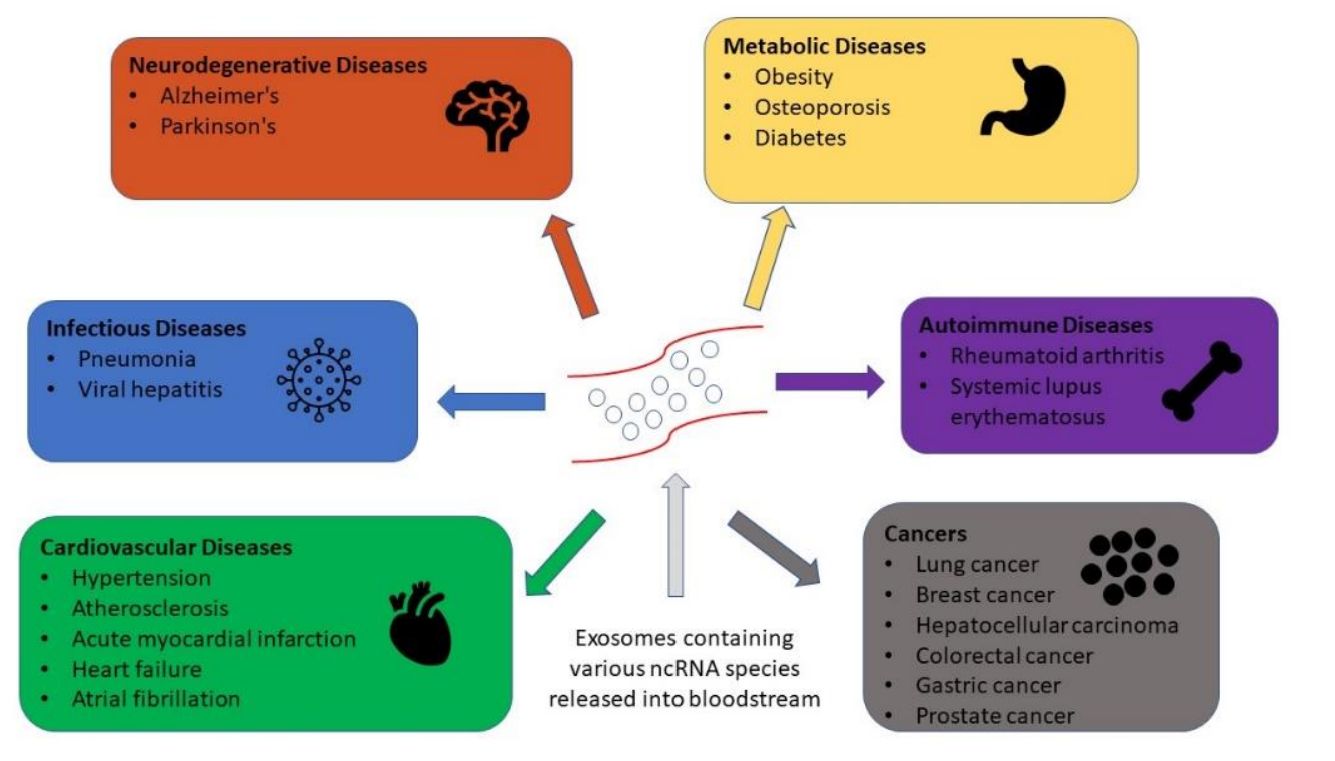
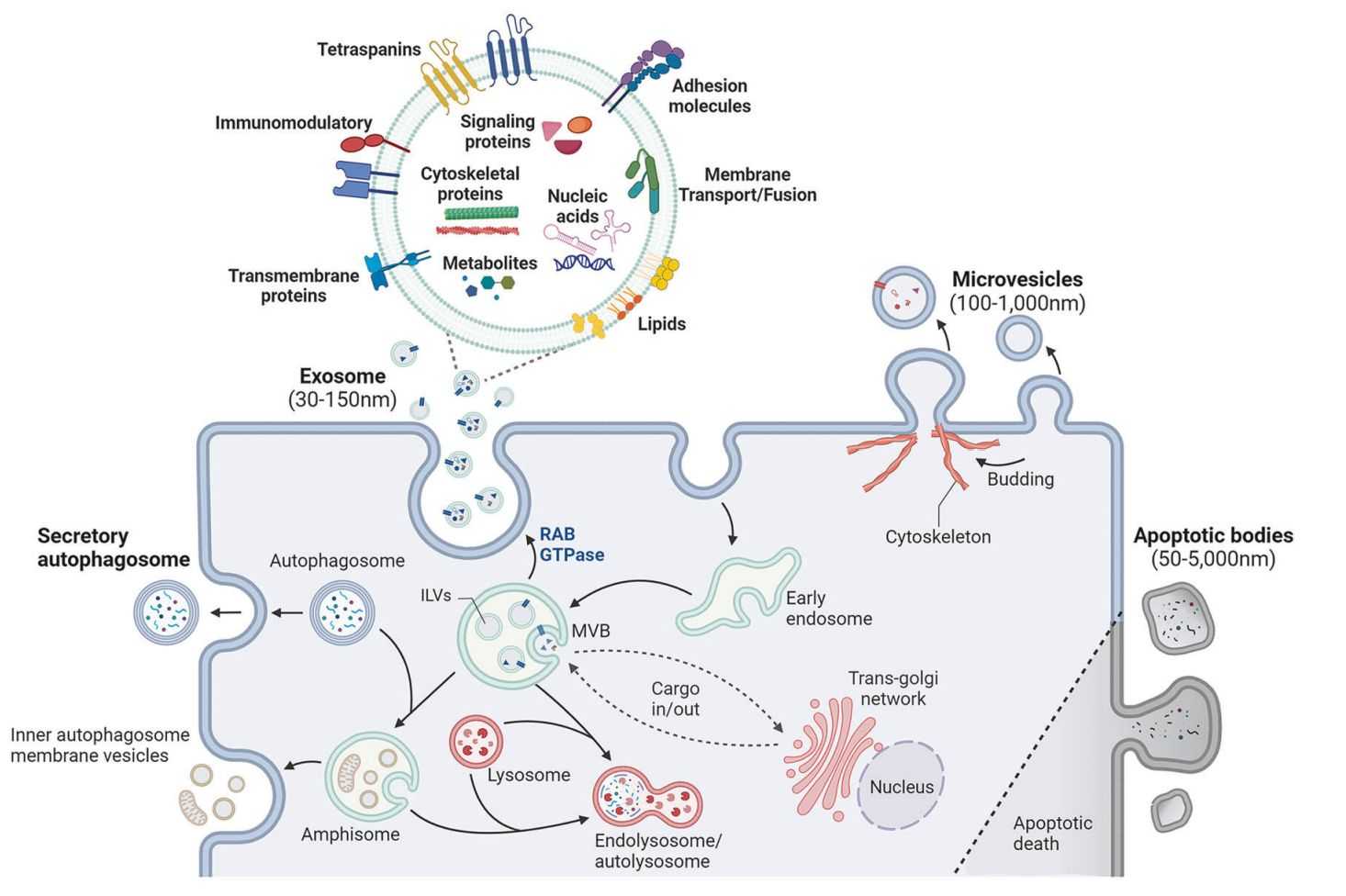
Exosome omics services profile the molecular cargo carried by exosomes to reveal what cells are doing in health and disease. Because exosomes circulate in biofluids and mirror their cells of origin, omics readouts can support biomarker discovery, patient stratification, and mechanism studies.
Each omics layer answers a different question. Exosome transcriptome profiling focuses on RNA content such as miRNA and mRNA to capture gene regulation signals. Exosome proteomics measures proteins to identify functional pathways, surface markers, and disease related changes. Exosome lipidomics characterizes lipid species that shape membrane properties and signaling. Exosome metabolomics profiles small molecules to reflect metabolic states and treatment response.
Multi omics integration combines these datasets to find consistent, cross validated signals and build pathway or network level interpretations.
Comprehensive Exosome Omics Solutions
Creative Biostructure offers an integrated portfolio of exosome omics services to help you decode exosomal cargo at multiple molecular levels. From discovery to interpretation, our workflows emphasize data quality, reproducibility, and biologically meaningful insights for biomarker research and translational studies.
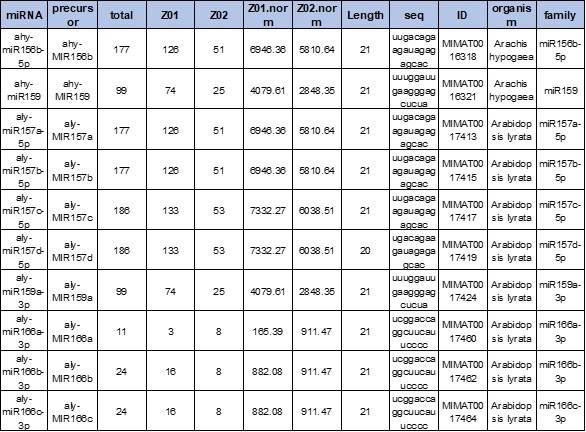
Uncover the complete exosomal RNA landscape, including mRNA, miRNA, lncRNA, and circRNA. We utilize high-sensitivity NGS protocols optimized for low-input samples to detect novel transcripts and analyze differential expression.
Achieve deep coverage of the exosomal proteome using high-resolution LC-MS/MS. Our services support Label-free, TMT, and DIA quantification strategies to identify surface markers, signaling proteins, and post-translational modifications.
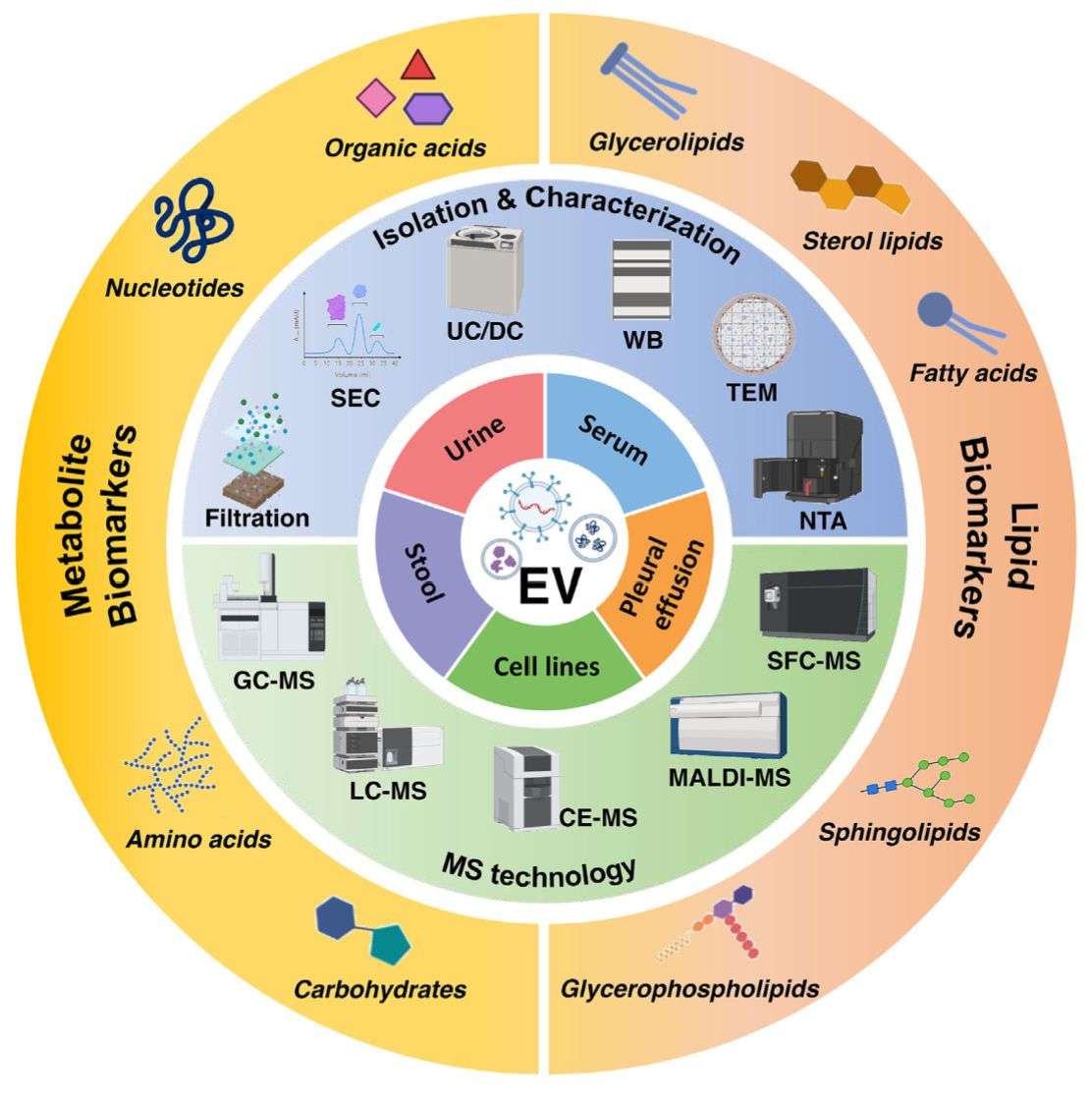
Characterize the lipid composition and membrane dynamics of exosomes. We perform targeted and untargeted profiling to identify lipid classes, fatty acid chains, and lipid-protein interactions crucial for vesicle stability and uptake.
Analyze the small-molecule metabolic signature of exosomes. Our metabolomics profiling provides a snapshot of the parent cell's physiological state, helping to identify metabolic reprogramming and potential diagnostic metabolites.
Synthesize data across molecular layers using advanced bioinformatics. We statistically correlate transcriptomic, proteomic, and metabolic datasets to map regulatory networks and identify robust, multi-dimensional biomarker signatures.
Explore Advanced Exosome Multi-Omics Technologies
Creative Biostructure applies fit for purpose omics platforms and rigorous quality control to generate reproducible, publication ready exosome datasets. We support discovery, targeted validation, and multi layer interpretation for biomarker and mechanism studies.
| Category | Techniques | Key Advantages | Best Use Cases |
|---|---|---|---|
| Transcriptomic Profiling (RNA) | Next-Generation Sequencing (NGS) | Captures full RNA diversity (mRNA, miRNA, lncRNA, circRNA) with high sensitivity for low-input samples. | Novel transcript discovery, differential expression analysis, and non-coding RNA regulatory studies. |
| Long-Read Sequencing (PacBio/Nanopore) | |||
| Single-Cell RNA Sequencing (scRNA-seq) | |||
| Proteomic Analysis (Protein) | Label-Free Quantification (LFQ) | Achieves deep proteome coverage and high quantitative accuracy without labeling bias. | Global protein profiling, post-translational modification (PTM) analysis, and biomarker identification. |
| TMT/iTRAQ Labeling | |||
| Data-Independent Acquisition (DIA) | |||
| Lipidomics & Metabolomics | UPLC-MS/MS (Targeted/Untargeted) | Provides precise structural identification and quantification of diverse lipid classes and polar metabolites. | Membrane dynamics research, metabolic reprogramming analysis, and lipid-protein interaction studies. |
| Shotgun Lipidomics | |||
| GC-MS | |||
| Bioinformatics & Integration | Multi-Omics Factor Analysis (MOFA) | Mathematically integrates heterogeneous datasets to reveal hidden sources of variation. | Systems biology modeling, multi-dimensional biomarker signature discovery, and pathway network construction. |
| Weighted Gene Co-expression Network Analysis (WGCNA) | |||
| Machine Learning Algorithms |
Applications of Exosome Omics and Multi-Omics Analysis
Precision Biomarker Discovery
- • Identifies robust multi-analyte signatures (RNA + Protein).
- • Enhances diagnostic sensitivity and specificity.
- • Distinguishes comorbidities through complex profiles.
Disease Mechanism Elucidation
- • Maps intricate regulatory networks and signaling pathways.
- • Links gene expression changes to metabolic outputs.
- • Uncovers novel pathogenesis in cancer and neurodegeneration.
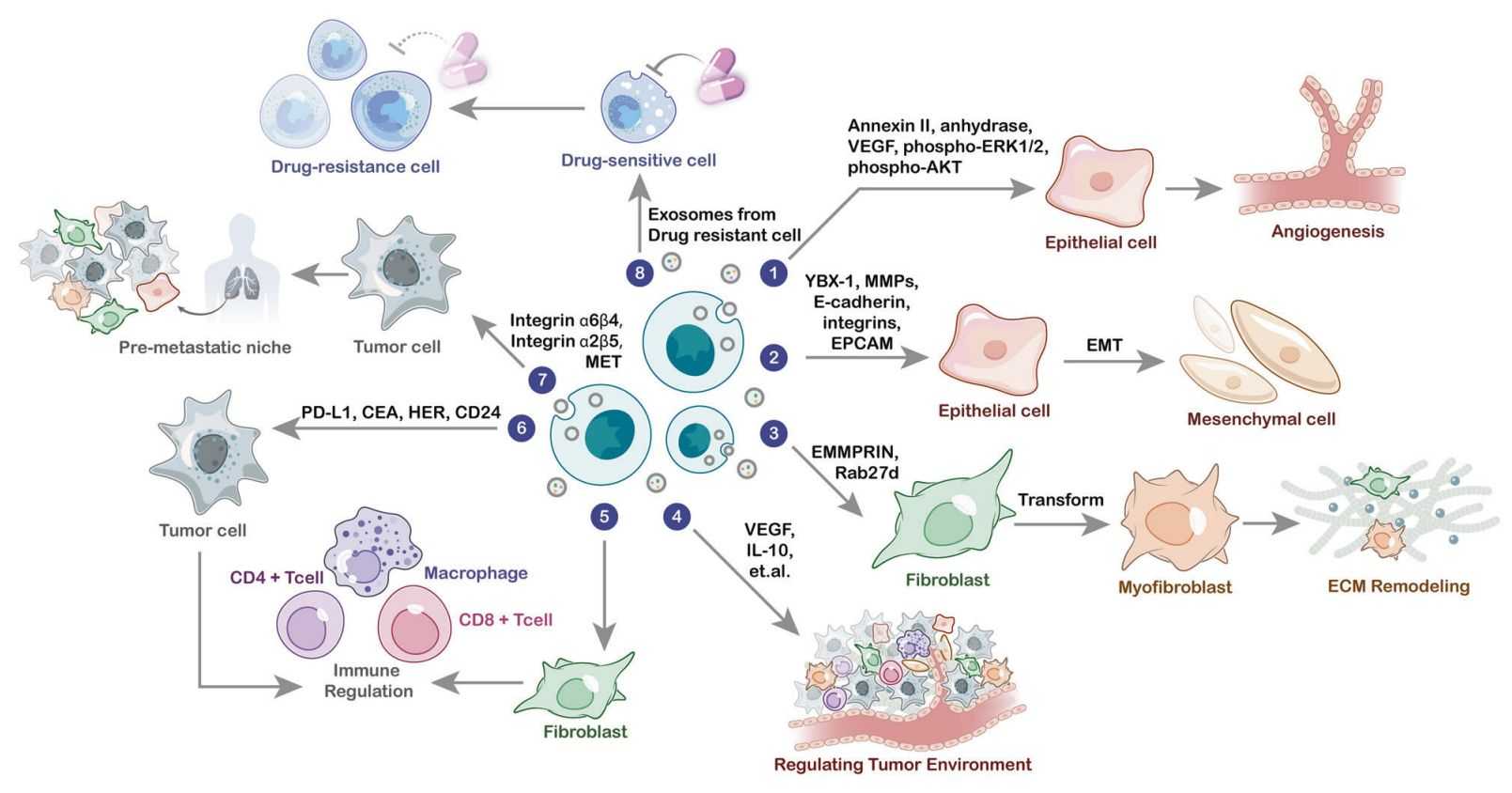
Therapeutic Target Identification
- • Reveals "druggable" nodes within disease networks.
- • Identifies mechanisms of drug resistance.
- • Validates therapeutic targets using systems biology.
Liquid Biopsy Development
- • Accelerates the development of non-invasive diagnostic tests.
- • Enables longitudinal monitoring of patient response.
- • Supports patient stratification for personalized medicine.
By integrating genomic, proteomic, and metabolic data, researchers can move beyond simple correlation to understanding causation. Creative Biostructure supports projects from initial multi-omics screening to rigorous validation and translation.
State-of-the-Art Multi-Omics Analytical Platforms
Creative Biostructure employs a suite of cutting-edge high-throughput technologies to drive multi-omics discovery. Our facility is equipped with Illumina NovaSeq and PacBio sequencing systems for comprehensive transcriptomics, alongside Thermo Scientific Orbitrap and Bruker timsTOF mass spectrometers for high-resolution proteomics, lipidomics, and metabolomics. Supported by our High-Performance Computing clusters for advanced bioinformatics integration, these platforms deliver exceptional data depth, sensitivity, and reproducibility for systems biology research.
Image Source: Illumina

Image Source: Thermo Fisher Scientific

Image Source: Bruker
Our Integrated Exosome Multi-Omics Workflow
At Creative Biostructure, our multi-omics workflow transforms complex samples into actionable insights. We use an optimized pipeline to preserve fragile cargo—RNA, proteins, lipids, and metabolites—ensuring high-quality data for biomarker discovery.
Sample Pre-processing and Quality Control
Perform rigorous quality checks and pre-treatment to remove cellular debris, ensuring optimal preservation of fragile RNA and metabolite markers.
Exosome Isolation and Cargo Extraction
Isolate high-purity exosomes using tailored methods (e.g., SEC) and employ specialized protocols for simultaneous extraction of RNA, proteins, and lipids to maximize yield.
High-Throughput Multi-Omics Profiling
Execute parallel data acquisition using NGS for transcriptomics and High-Resolution LC-MS/MS for proteomics, lipidomics, and metabolomics.
Integrated Bioinformatics Analysis
Perform statistical integration of multi-omics datasets to uncover regulatory networks and identify robust composite biomarker signatures.
Biological Interpretation and Reporting
Deliver publication-ready reports with interactive visualizations, offering deep biological interpretation that connects data to functional mechanisms.
Creative Biostructure offers flexible service options: choose individual omics profiling (e.g., RNA-seq only) or a fully integrated multi-omics package tailored to your specific research questions.
Starting a Project with Creative Biostructure
Sample Requirements
- • Minimum Volume: Varies by omics combination; typically 3-5 mL for biofluids (plasma/serum) or >50 mL for cell culture supernatants to ensure sufficient yield for simultaneous RNA/Protein/Lipid extraction.
- • Acceptable Sample Types: Cell culture media, plasma (EDTA preferred), serum, urine, CSF, or pre-purified exosome pellets.
- • Storage and Transport: Samples must be snap-frozen in liquid nitrogen and shipped on dry ice to prevent RNA degradation and metabolic changes.
- • Critical Handling: Strictly avoid repeated freeze-thaw cycles and hemolysis, as these significantly alter the exosomal multi-omics profile.
For complex multi-omics designs, please contact our technical team to discuss specific input requirements based on your target sensitivity.
Deliverables
- • Comprehensive Project Report: Detailed experimental design, library preparation/sample prep methods, and rigorous QC metrics (e.g., RNA RIN, peptide coverage).
- • Raw & Processed Data: Full access to raw data files (FASTQ for NGS, .raw for MS) and normalized data matrices (Excel/CSV).
- • Bioinformatics Analysis: Differential expression analysis, pathway enrichment (GO/KEGG), and multi-omics integration networks.
- • Visualizations: Publication-ready figures including Heatmaps, Volcano plots, PCA plots, and Circos plots.
Our data packages provide a holistic systems-biology view, equipping you with the statistical evidence needed for high-impact publications and biomarker validation.
Featured Exosome Omics Case Studies
At Creative Biostructure, we bridge the gap between raw data and biological insight. Discover how our integrated multi-omics strategies empower researchers to decipher complex disease mechanisms and secure high-impact publications.
Why Choose Our Exosome Omics Analysis Services
Creative Biostructure leverages cutting-edge NGS and Mass Spectrometry platforms to deliver seamless multi-dimensional data integration, turning complex exosomal cargo into clear biological insights.
True Multi-Omics Integration
Seamlessly combine Transcriptomics, Proteomics, and Lipidomics under one roof to ensure datasets are perfectly aligned for robust correlation analysis.
Superior Sensitivity for Low-Input Samples
Optimized protocols allow deep profiling from limited biofluids, maximizing information retrieval and data depth without compromising sample integrity.
Advanced Systems Biology Analysis
We utilize advanced statistical models to map regulatory networks, linking gene expression to metabolic output to reveal underlying disease mechanisms.
Tailored Experimental Design
Flexible designs from discovery to validation, with expert consultation to ensure strategies align precisely with your research goals.

What Our Customers Say
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.



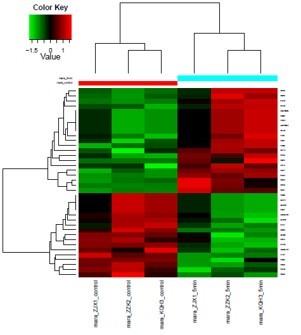
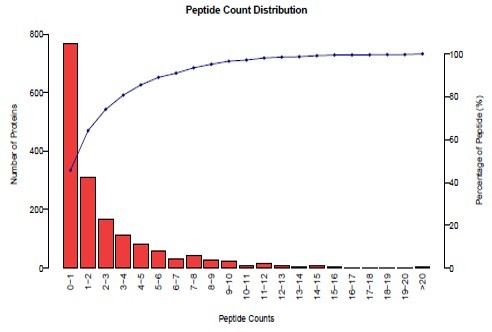
" We used their Label-free proteomics to map exosome surface markers. The sensitivity for low-abundance proteins was impressive and the raw data was incredibly clean. The final report was comprehensive and ready for our publication."
– Sarah Jenkins, Senior Scientist, Translational Medicine
" Their small RNA-seq workflow handled our low-input plasma samples perfectly. The bioinformatics team's pathway analysis helped us pinpoint valid miRNA biomarkers."
– Dr. Aris Thorne, Principal Investigator, Cancer Biology
" Accurate lipid profiling was vital for our engineered exosome stability. Their targeted lipidomics platform provided the quantitative resolution we lacked internally."
– Mark Davis, Lead Bioengineer, Drug Delivery Systems