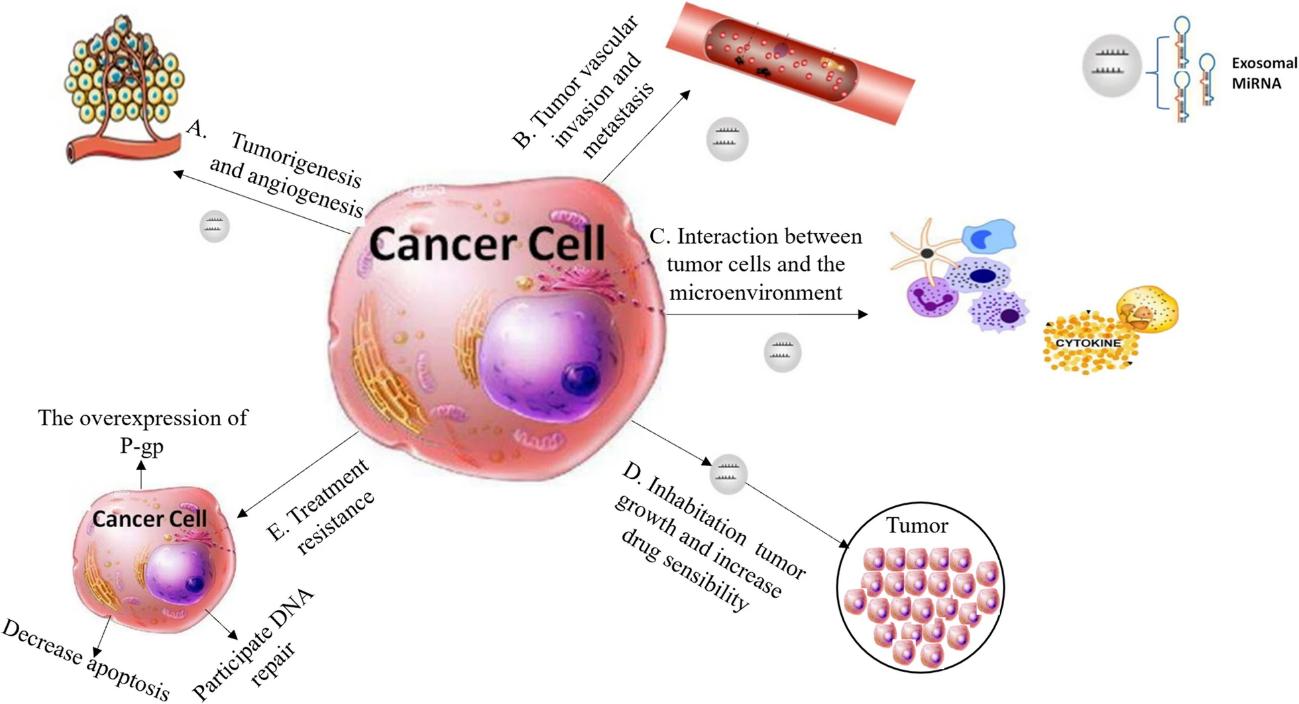
Exosomal Whole Transcriptome Sequencing Service
Exosomes, as nanoscale vesicles secreted by cells, carry a large number of RNA molecules to facilitate intercellular communication. Their transcriptomic characteristics provide crucial clues for disease diagnosis and mechanism research. Creative Biostructure offers comprehensive exosome transcriptome sequencing services. Our services serve as a powerful tool for disease biomarker screening, global gene regulation studies, exploration of disease mechanisms, and development of novel diagnostic strategies, providing robust data support for clients' basic research and clinical translational studies.
What is Exosomal Whole Transcriptome Sequencing and Why is it Used?
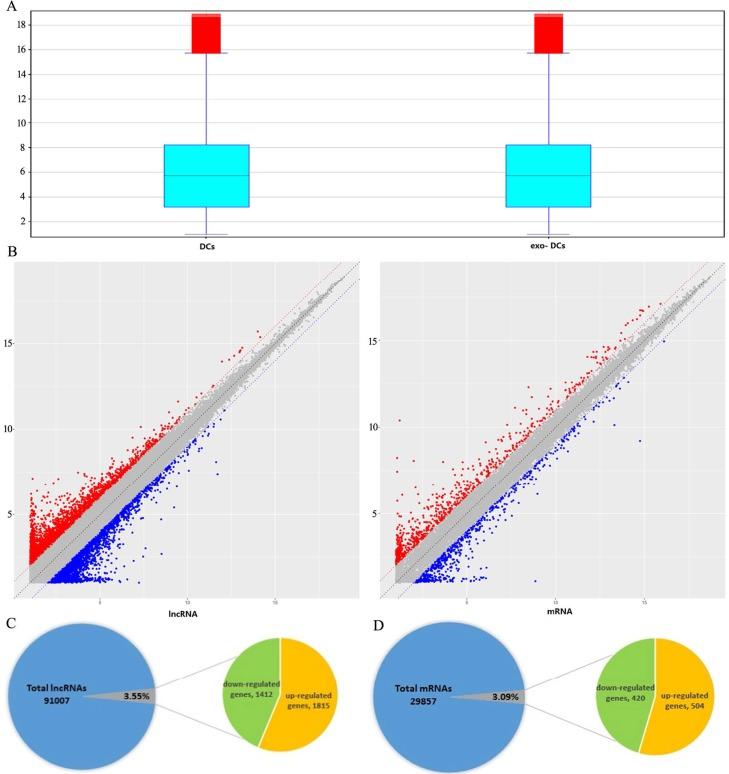
Exosomes carry important information such as proteins, lipids, and RNAs derived from their parent cells. Whole-transcriptome sequencing analysis of long RNAs in exosomes enables the detection of data from tens of thousands of long RNAs, including mRNA, lncRNA, and circRNA, in a single sequencing experiment. With its large data volume and comprehensive information, exosome whole-transcriptome sequencing serves as a critical tool for identifying specific disease diagnostic biomarkers and conducting multi-omics analyses. Furthermore, since over 70% of the long RNAs carried by exosomes are mRNAs, the results of exosome whole-transcriptome sequencing not only allow for in-depth gene functional annotation but also facilitate the analysis of potential cell types of origin and their proportional contributions.
Our Exosomal Whole Transcriptome Sequencing Services
Creative Biostructure offers a comprehensive exosome whole transcriptome sequencing service. This system fully integrates low-input nucleic acid library construction technology, NGS technology, and novel bioinformatic sequence alignment algorithms. It enables complete capture and sequencing of various RNAs, including mRNA, lncRNA, and circRNA, from plasma exosomes. With just 1 mL of plasma, the service reliably detects over 10,000 long RNA transcripts.
| RNA Types | Fundamental Analysis | Advanced Analysis | |
|---|---|---|---|
| mRNA |
|
|
|
| IncRNA |
|
|
|
| circRNA |
|
|
|
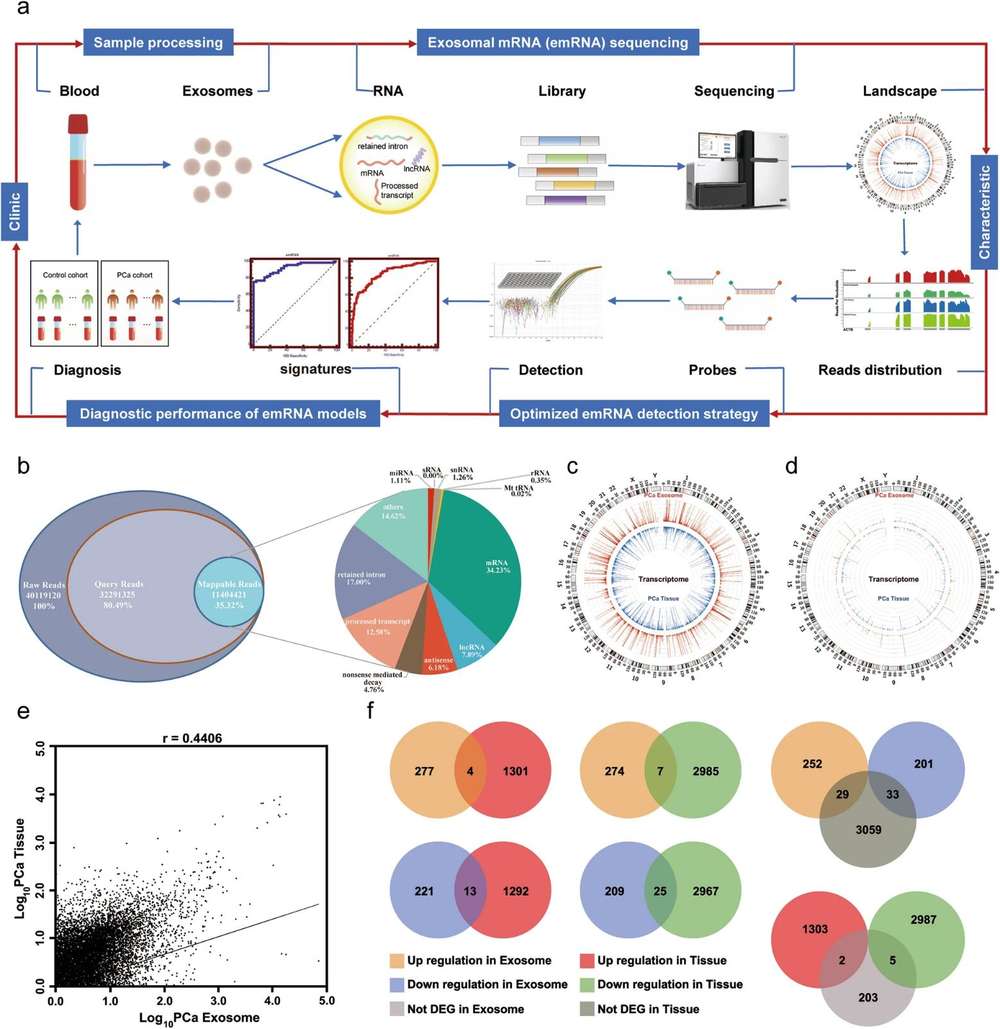
Exosomal Whole Transcriptome Sequencing Analysis Workflow
Our exosomal whole transcriptome sequencing service employs standardized procedures to ensure stable and reproducible results across various sample types.
Exosome Isolation
Exosomes are isolated from biological fluids or cell culture supernatants using techniques such as ultracentrifugation, size-exclusion chromatography, immunoaffinity capture, or microfluidic technology, followed by strict quality control of particle size and purity.
RNA Extraction and Quality Control
Total exosomal RNA extraction is followed by comprehensive assessment of RNA integrity and concentration on a high-sensitivity platform.
Library Construction
After RNA fragmentation, cDNA is synthesized through reverse transcription, followed by the ligation of sequencing adapters and PCR amplification under optimized conditions to maximize transcript diversity and minimize technical bias.
Sequencing
High-throughput sequencing is performed using the Illumina platform, generating high-quality reads, with sequencing depth flexibly adjustable depending on research needs.
Bioinformatics Analysis
Data are processed through a rigorous pipeline beginning with quality control, followed by reference sequence alignment, transcript quantification, differential expression analysis, and concluding with functional annotation. A suite of extended analyses is also offered, such as pathway enrichment, novel transcript discovery, and biomarker mining.
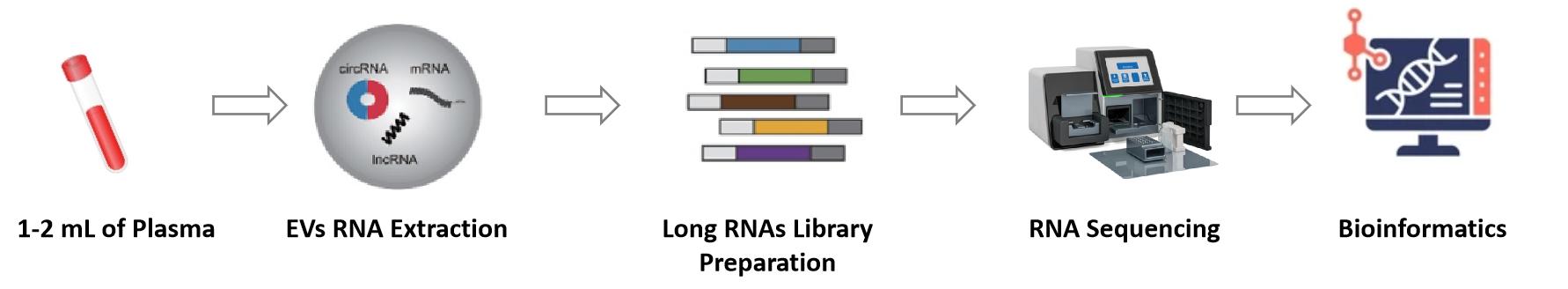 Figure 1. Workflow for exosomal whole transcriptome sequencing and analysis services. (Creative Biostructure)
Figure 1. Workflow for exosomal whole transcriptome sequencing and analysis services. (Creative Biostructure)
Advantages of Our Exosomal Whole Transcriptome Sequencing Services
- One-stop Service System
Clients only need to provide plasma samples, and our experienced scientists will manage the entire service process for them, encompassing sample preparation, library construction, sequencing, and data analysis.
- Extensive RNA Analysis
Our platform detects a full range of RNA types, such as mRNAs, lncRNAs, and circRNAs, facilitating a thorough investigation of exosomal transcriptomes.
- Professional Bioinformatics Analysis
Creative Biostructure boasts a robust bioinformatics team capable of meeting clients' diverse and in-depth data analysis requirements.
Case Study
Case: Integrating scRNA-seq of PBMCs and Exosomal Transcriptomics to Uncover Cellular and Genetic Landscapes in COPD
Background
This study performed whole-transcriptome sequencing on plasma exosomes from patients with chronic obstructive pulmonary disease (COPD) and healthy controls, and conducted single-cell RNA sequencing on peripheral blood mononuclear cells (PBMCs). Ultimately, competitive endogenous RNA (ceRNA) networks and protein-protein interaction (PPI) networks were constructed to elucidate the interaction mechanisms between PBMCs and exosomes in COPD patients.
Methods
- Exosome isolation: The ethylenediaminetetraacetic acid-anticoagulated blood samples were centrifuged at 10,000 × g for 15 min at 4 °C to remove debris.
- Exosome characterization: Transmission electron microscopy (TEM) was used to examine the morphology of the isolated exosomes.
- RNA extraction, library construction, and RNA sequencing: Prior to sequencing, RNA was extracted from exosomes using TRIzol reagent. The purified RNA was then used to construct a cDNA library using an MGIEasy RNA Directional Library Prep Kit (MGI). The constructed library was sequenced on an Illumina DNBSEQ platform, the raw reads generated were filtered using SOAPnuke software to obtain clean reads, and were saved in FASTQ format.
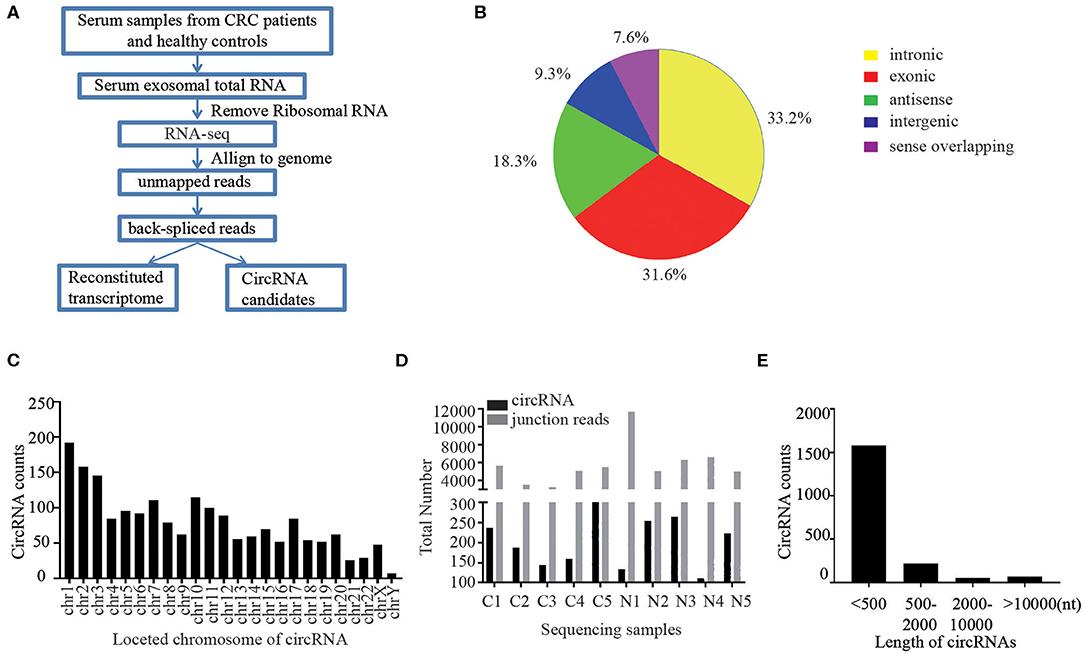
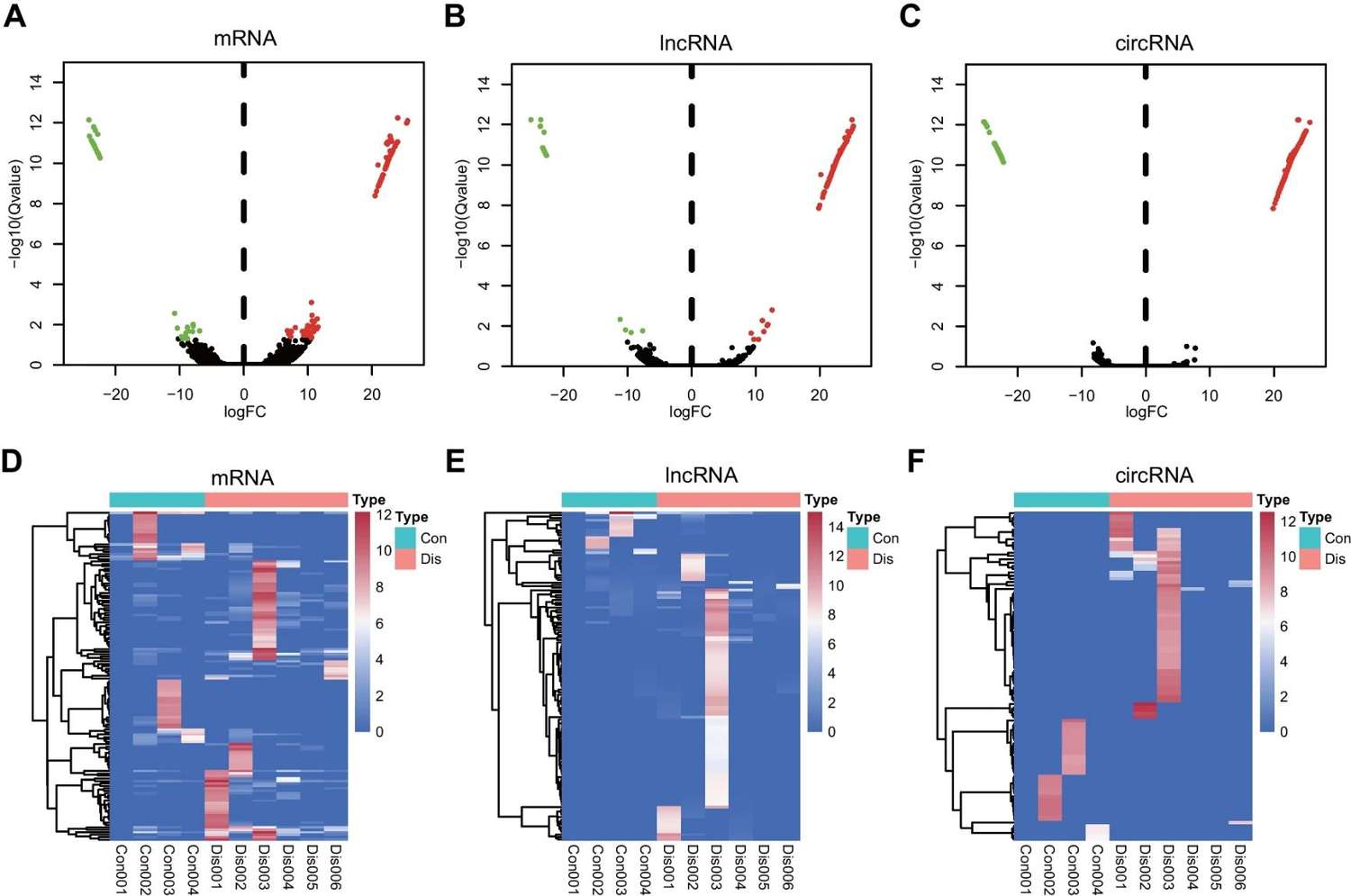 Figure 2 Identification of exosomal differentially expressed mRNAs, lncRNAs, and circRNAs in COPD. (Pei Y, et al., 2022)
Figure 2 Identification of exosomal differentially expressed mRNAs, lncRNAs, and circRNAs in COPD. (Pei Y, et al., 2022)
- Competing endogenous RNA (ceRNA) network construction: Bioinformatic databases were applied to construct ceRNA network.
- Single-cell preparation and sequencing: PBMCs were isolated from another blood samples of Dis6 and Con4 using the Ficoll-Paque Plus reagent (GE Healthcare). The PBMC suspensions were then loaded onto a haemocytometer for cell counting and viability examination.
Conclusion
A total of 135 mRNAs, 132 lncRNAs, and 359 circRNAs were identified in exosomes, which exhibited differential expression in six patients with chronic obstructive pulmonary disease compared to four healthy controls. Functional enrichment analysis revealed that many of these differentially expressed RNAs participate in immune responses, including defense against viral infection and cytokine-cytokine receptor interactions.
Creative Biostructure offers comprehensive services spanning sample consultation, protocol optimization, and result interpretation. We are committed to providing professional and efficient support to accelerate your research journey from basic studies to biomarker discovery and drug target screening. Contact us for more details, we are dedicated to serving you.
Reference
- Pei Y, Wei Y, Peng B, et al. Combining single-cell RNA sequencing of peripheral blood mononuclear cells and exosomal transcriptome to reveal the cellular and genetic profiles in COPD. Respiratory Research. 2022, 23(1): 260.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.