Exosome Lipidomics and Metabolomics Profiling Service
Exosomes have the highest lipid-to-protein ratio among extracellular vesicles, with unique lipid/metabolite profiles that reflect cellular states and regulate recipient cell function (e.g., cancer cell metabolism reprogramming). Creative Biostructure employs UPLC-MS/MS and FTMS technologies to deliver precise qualitative/quantitative data, supporting biomarker identification, cancer research, and exosome biology exploration. Actionable results are delivered within 4-6 weeks, including comprehensive differential analysis and expert support.
Why Conduct Exosome Lipidomics and Metabolomics Services?
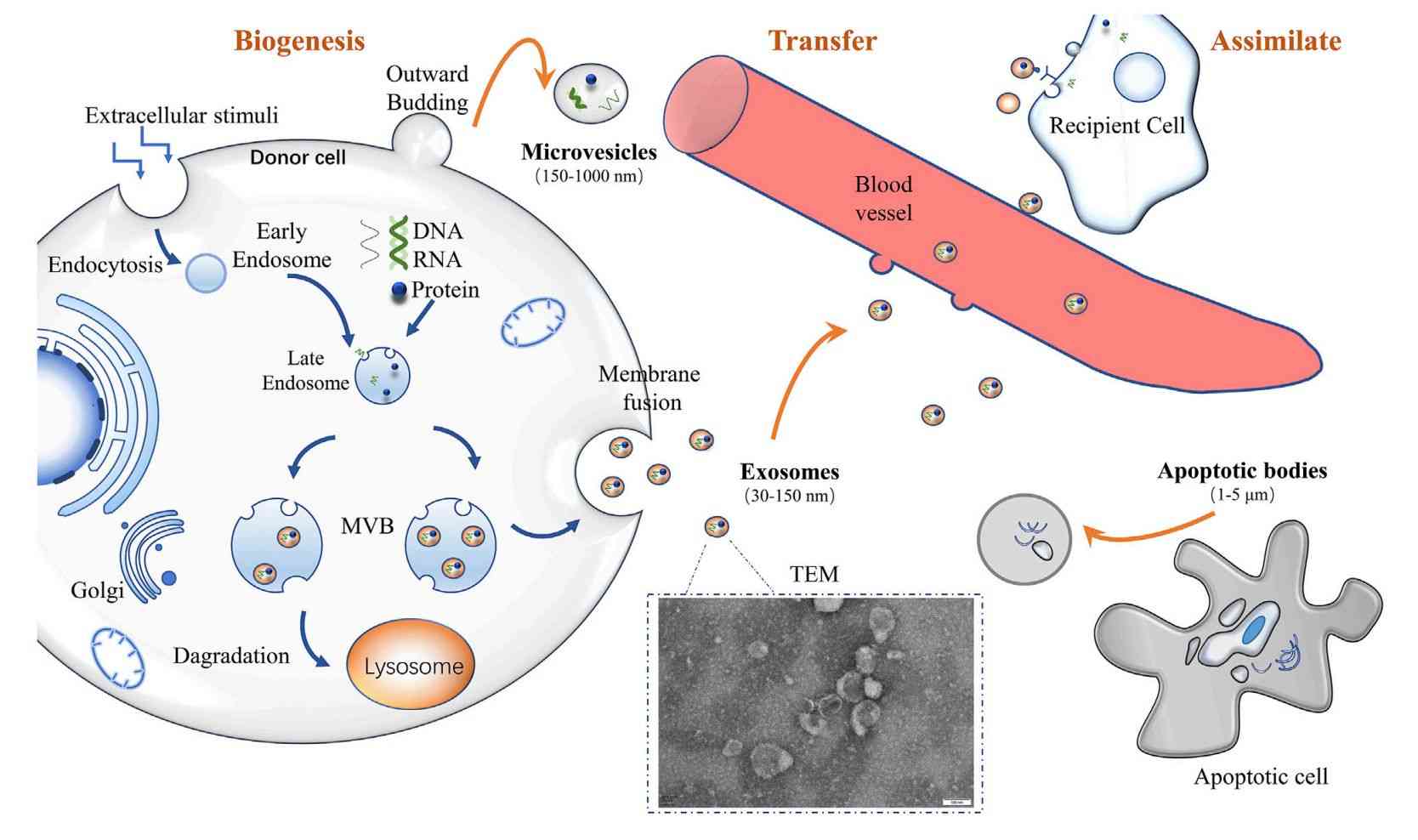
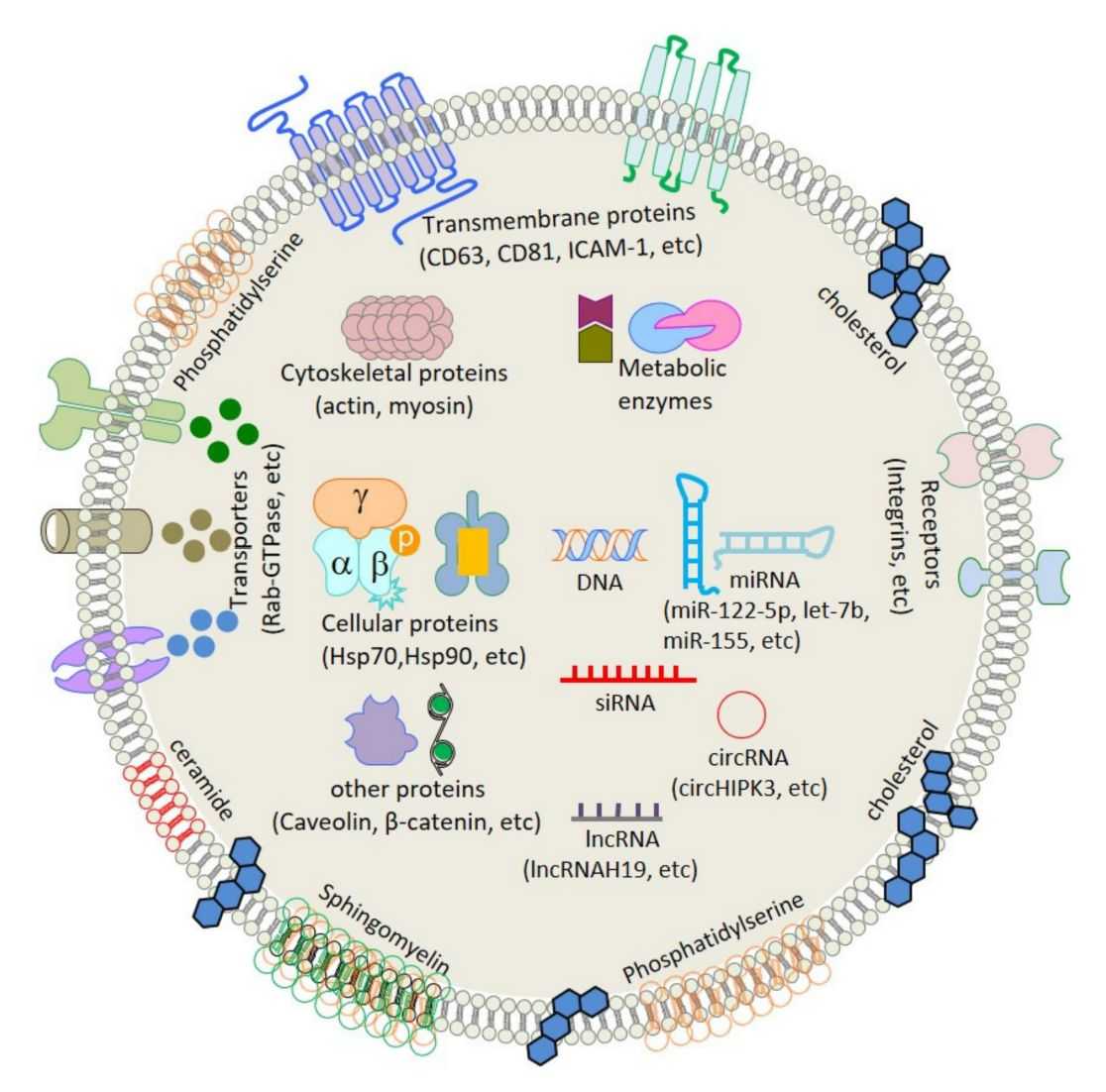
Exosomes are essential for intercellular communication. Their lipid and metabolite profiles directly reflect cellular function. Lipids form the exosomal membrane and guide its delivery, while metabolites reveal changes in the cellular environment. Together, they capture crucial differences between health and disease.
Comprehensive profiling of exosomes identifies the most active signaling molecules in cell-to-cell communication. This method detects changes in key pathways, including cell membrane structure, energy metabolism, and signal transduction during disease development. It can uncover early functional biomarkers that conventional omics approaches often miss. In cancer research, neurodegeneration, or drug response studies, exosomal lipidomics and metabolomics provide direct and dynamic functional insights.
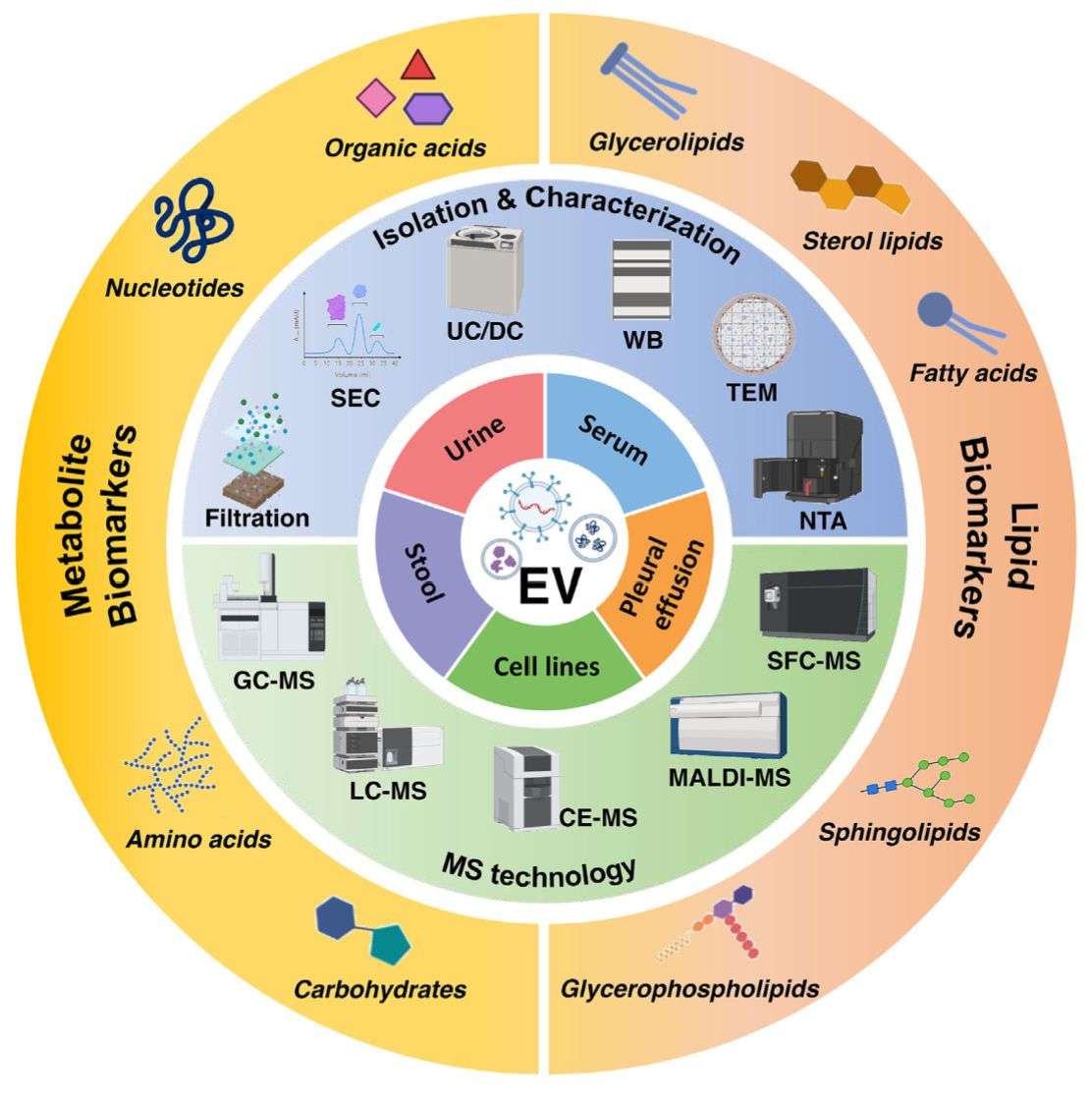
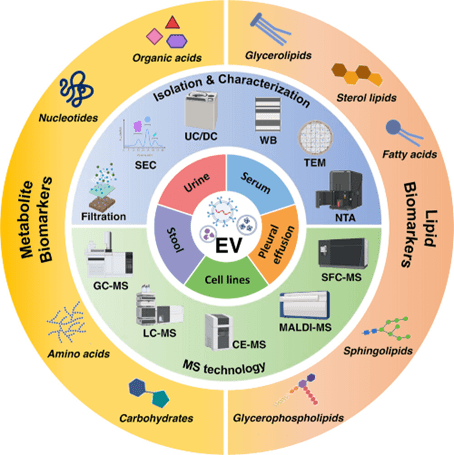 Figure 1. Advanced technologies for MS-based metabolomics and lipidomics of EVs. (Bai, L. et al., 2023)
Figure 1. Advanced technologies for MS-based metabolomics and lipidomics of EVs. (Bai, L. et al., 2023)
Our Exosome Lipidomics and Metabolomics Profiling Service
Our service provides comprehensive, high-resolution analysis of the molecular landscape within exosomes. By simultaneously profiling lipid and metabolite compositions, we deliver functional insights into intercellular communication, disease mechanisms, and therapeutic responses. We utilize high-sensitivity LC-MS/MS systems with optimized chromatographic separation for accurate identification and quantification of hundreds of lipid and metabolite species across multiple classes.
Key Features of Our Service:
| Services | Description |
|---|---|
| Exosome QC | Combine lipidomics and metabolomics in a single workflow to capture complementary functional data. |
| Dual-Omics Integration | We validate exosome purity using NTA, TEM, and Western Blot before analysis. |
| Targeted + Untargeted Strategies | Choose from precise targeted quantification or broad discovery-driven profiling. |
| Multi-Omics Data Correlation | Correlate lipid/metabolite profiles with transcriptomic or proteomic datasets. |
Technical Platform of Our Exosome Lipidomics and Metabolomics Services
We leverage cutting-edge mass spectrometry (MS) and separation technologies to ensure sensitivity, specificity, and reproducibility.
| Technical Platform | Description |
|---|---|
| UPLC-MS/MS | For targeted quantification of polar/non-polar lipids (e.g., glycerophospholipids, sphingolipids) and metabolites (e.g., amino acids, TCA-cycle intermediates). |
| FTMS | Enables high-resolution analysis of low-abundance metabolites, reducing matrix interference. |
| Bligh-Dyer Method | Gold-standard lipid extraction to preserve lipid integrity and avoid oxidation. |
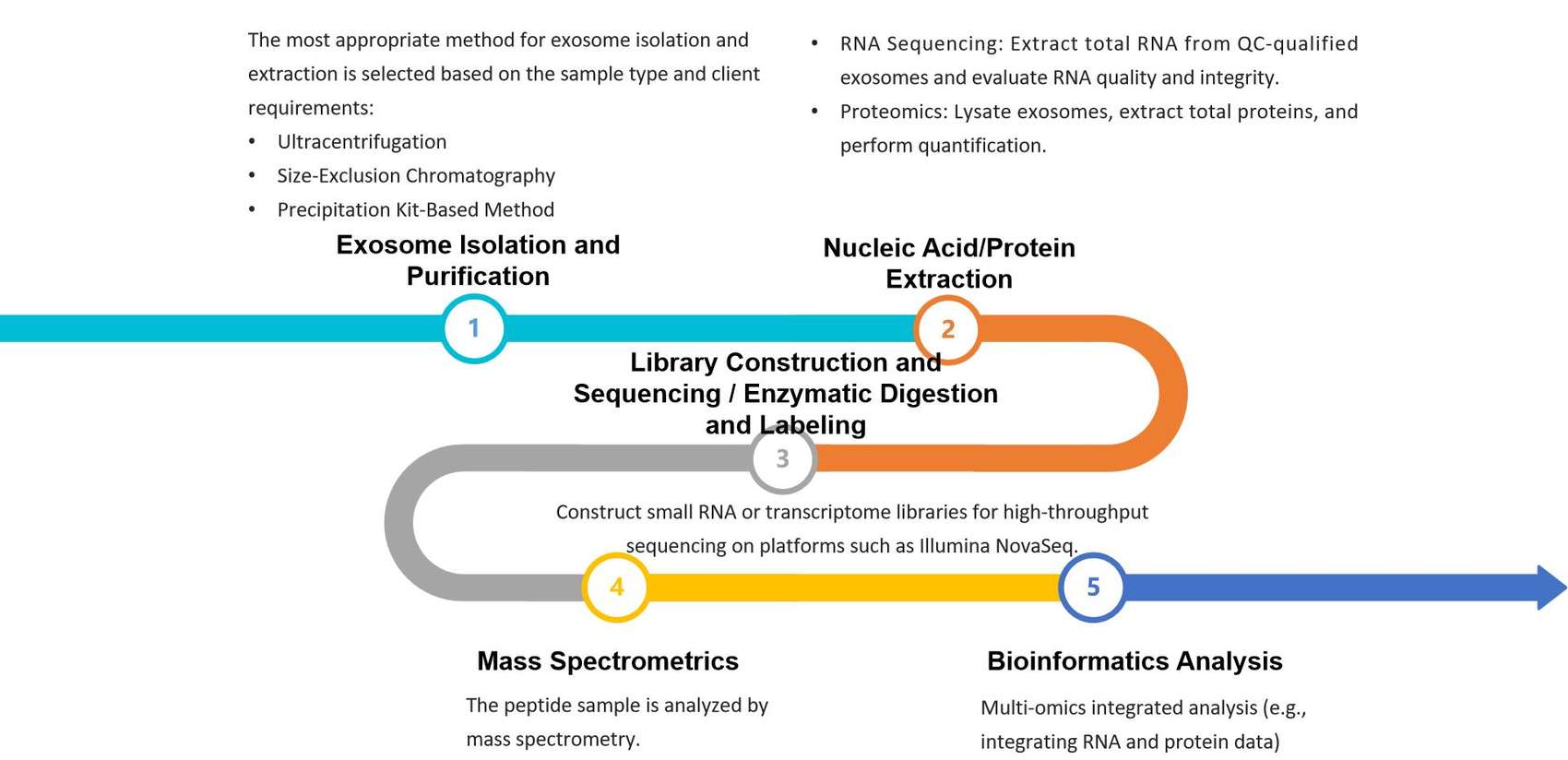
Workflow of Our Exosome Lipidomics and Metabolomics Analysis
Our end-to-end workflow is optimized to minimize sample loss and maximize data quality.
Sample Receipt & QC
Verify sample integrity (volume, storage condition) and assign project IDs.
Exosome Purification
Employ ultracentrifugation (the gold standard method) or immunoaffinity capture (for target-specific exosomes) to isolate pure exosomes and remove contaminating proteins and free nucleic acids.
Lipid/Metabolite Extraction
Separate polar (e.g., amino acids, organic acids) and non-polar (e.g., lipids) fractions via solvent partitioning.
MS Detection
Run targeted scans for pre-defined lipid/metabolite panels, with isotope internal standards for quantification.
Data Analysis
Conduct differential analysis (PCA, volcano plots), GO/KEGG pathway enrichment, and putative identification (via m/z ratio and retention time).
Report Delivery
Share results within 4-6 weeks (faster turnaround available for urgent projects).
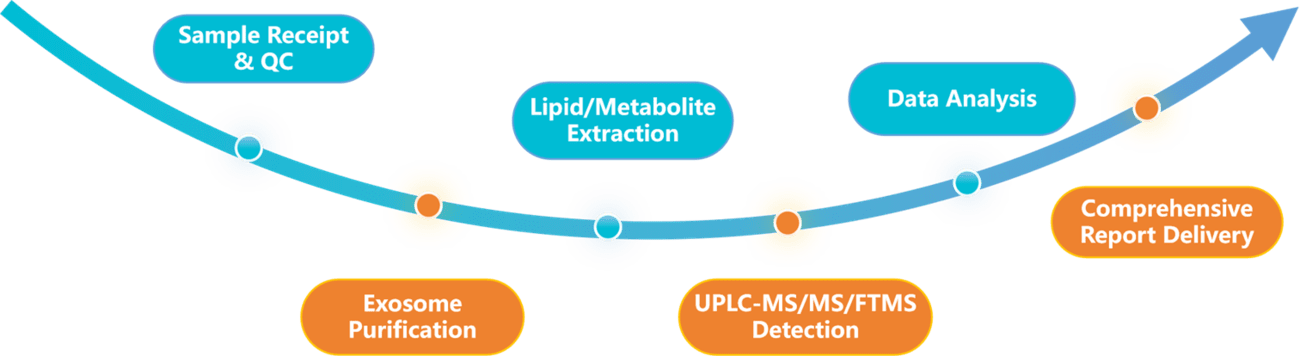 Figure 2. Comprehensive workflow for combined exosome lipidomics and metabolomics. (Creative Biostructure)
Figure 2. Comprehensive workflow for combined exosome lipidomics and metabolomics. (Creative Biostructure)
Sample Requirements
We accept a wide range of sample types and provide clear guidelines to ensure sample quality.
| Sample Type | Minimum Volume | Storage Condition |
|---|---|---|
| Serum/Plasma | 0.5-1 mL | -80°C, no repeated freeze-thaw |
| Cell Supernatant | 5-10 mL | -80°C, centrifuged to remove cells |
| Urine/Cerebrospinal Fluid | 3-5 mL | -80°C, preserved with protease inhibitor |
| Purified Exosomes | 100 μg (protein content) | -80°C |
Why Choose Our Service?
- High Specificity: Targeted analysis reduces "noise" compared to untargeted metabolomics, focusing on biologically relevant lipids/metabolites.
- Fast Turnaround: 4-6 weeks without compromising data quality.
- Expert Support: Dedicated scientists provide 1:1 consultation on experimental design and data interpretation.
- Reproducibility: Strict QC (technical replicates, standard curves) ensures consistent results across batches.
Case Study
Case: Plasma exosomal lipidomics identifies early-detection biomarkers for HCC in cirrhotic patients
Background
Novel biomarkers for HCC surveillance in cirrhotic patients are urgently needed. Exosomes and their lipid content in particular represent potentially valuable noninvasive diagnostic biomarkers. In this study, exosomes were isolated from plasma samples of 72 patients with liver cirrhosis and subjected to lipidomic analysis.
Research Highlights
- Exosomes and unfractionated plasma were processed for untargeted lipidomics using ultra–high-resolution mass spectrometry. A total of 2,864 lipid species, belonging to 52 classes, were identified.
- Both exosome fractionation and HCC diagnosis had significant impact on the lipid profiles. Ten lipid classes were enriched in HCC exosomes compared with non-HCC exosomes. Dilysocardiolipins were detected in 35% of the HCC exosomes but in none of the non-HCC exosomes (P < 0.001).
Conclusion
This study identifies lipids in circulating exosomes, that could serve as biomarkers for the early detection of hepatocellular carcinoma as well as altered pathways in exosomes that may contribute to tumor development and progression.
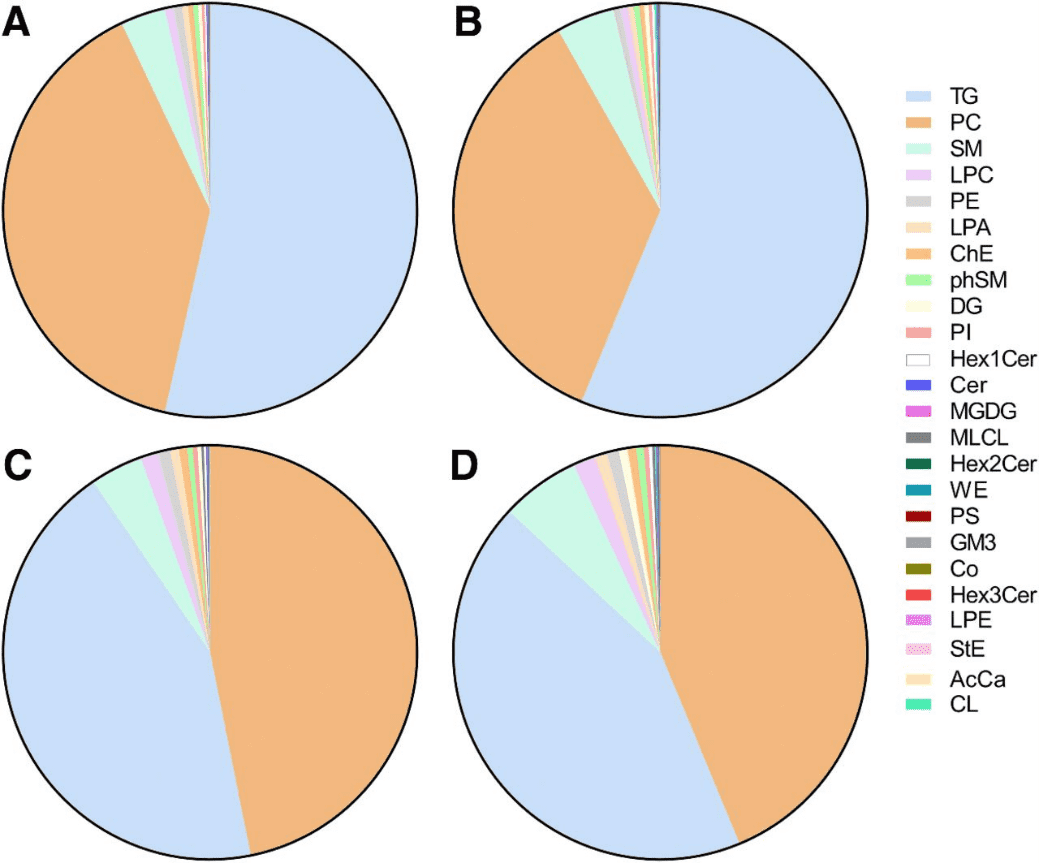 Figure 3. Distribution of lipid classes in plasma and exosomes from HCC and non-HCC patients. (Sanchez, J. I., et al., 2021)
Figure 3. Distribution of lipid classes in plasma and exosomes from HCC and non-HCC patients. (Sanchez, J. I., et al., 2021)
Creative Biostructure's Exosome Lipidomics and Metabolomics Profiling Service delivers integrated, precise dual-omics insights-unlocking lipid and metabolite interactions in exosomes to fuel your disease research, biomarker discovery, or therapeutic development. Leverage our cutting-edge platforms, optimized workflows, and expert data interpretation. Contact us today to share your project goals and partner with specialists for reliable, research-accelerating dual-profiling results.
References
- Sanchez, J. I., et al. Lipidomic profiles of plasma exosomes identify candidate biomarkers for early detection of hepatocellular carcinoma in patients with cirrhosis. Cancer Prevention Research. 14.10 (2021): 955-962.
- Bai, L., et al. Mass spectrometry‐based extracellular vesicle micromolecule detection in cancer biomarker discovery: an overview of metabolomics and lipidomics. View. 4.5 (2023): 20220086.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.