Exosome Proteomics Services
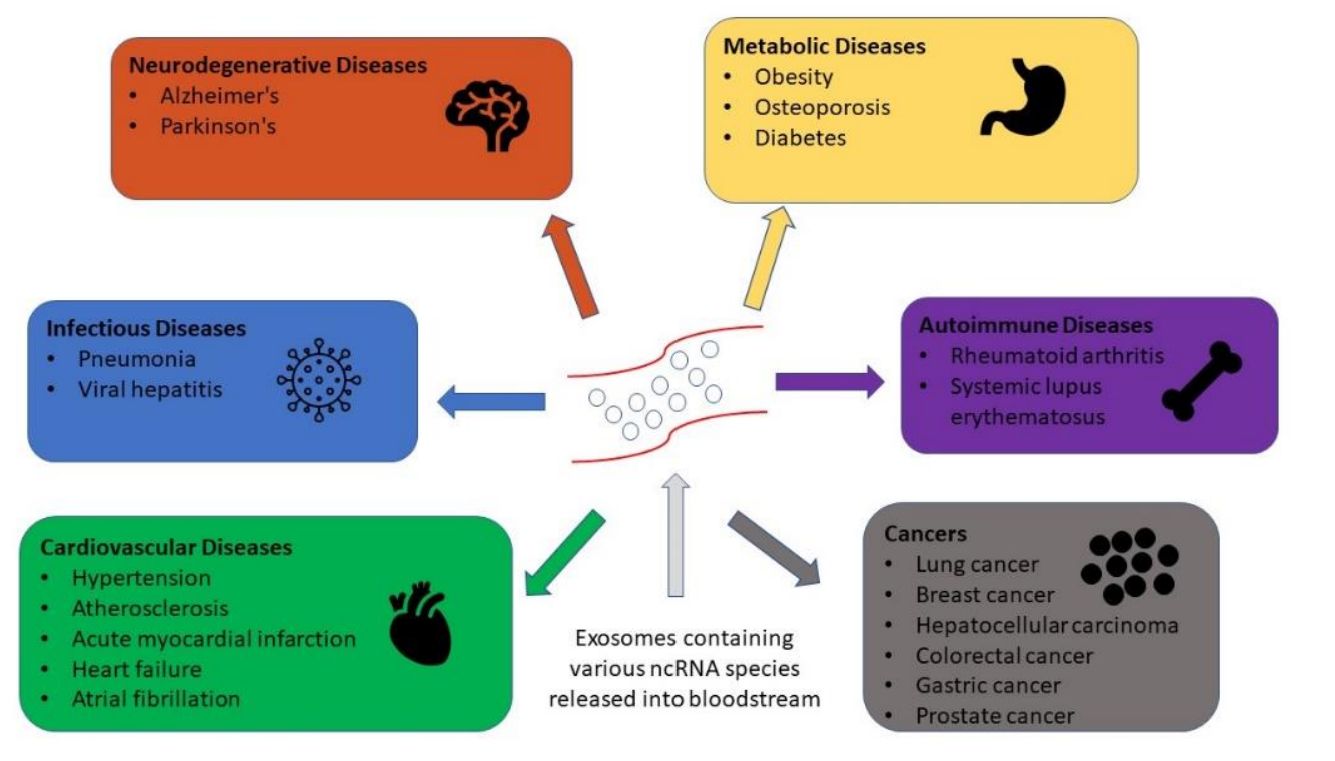
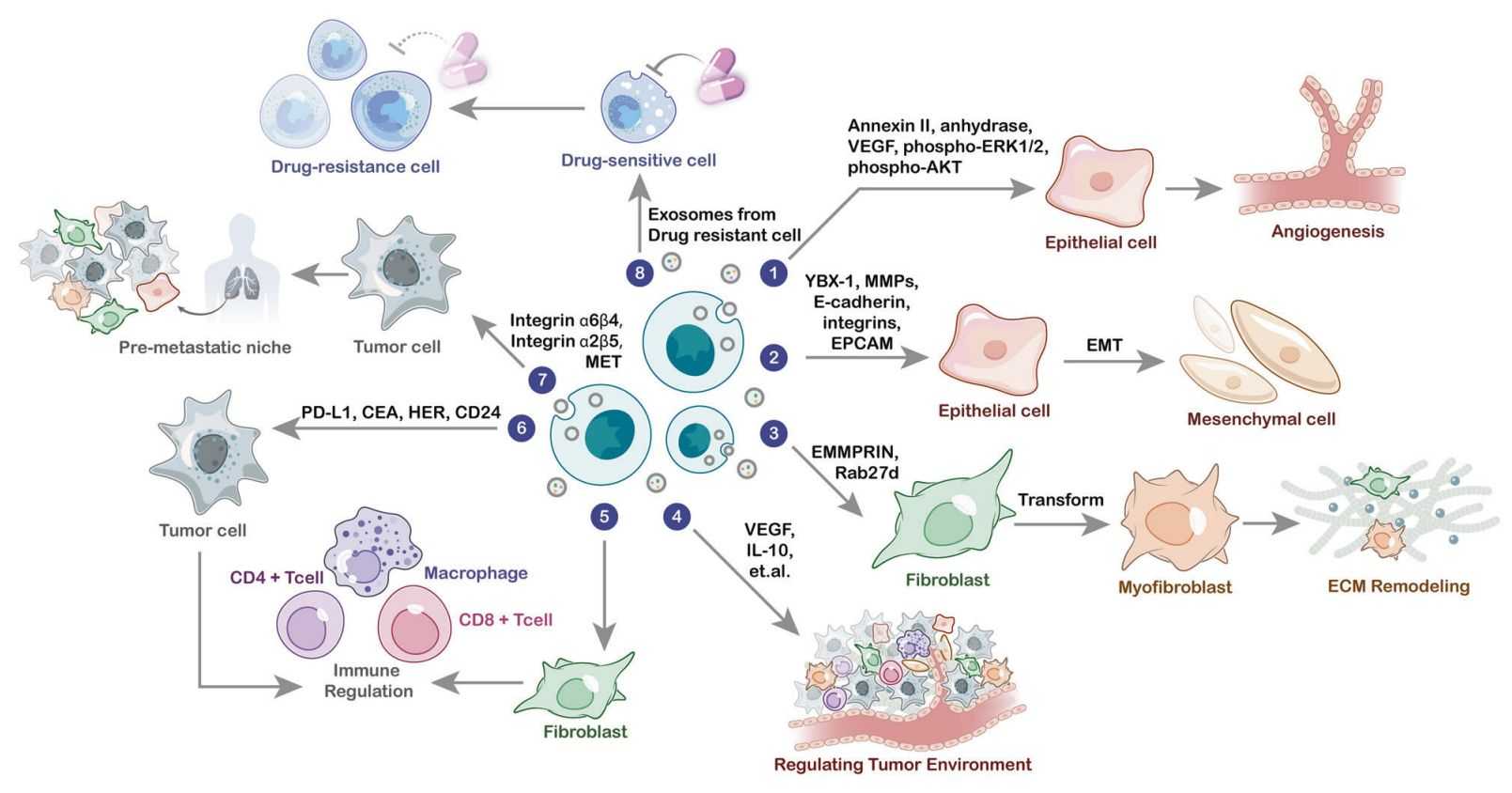
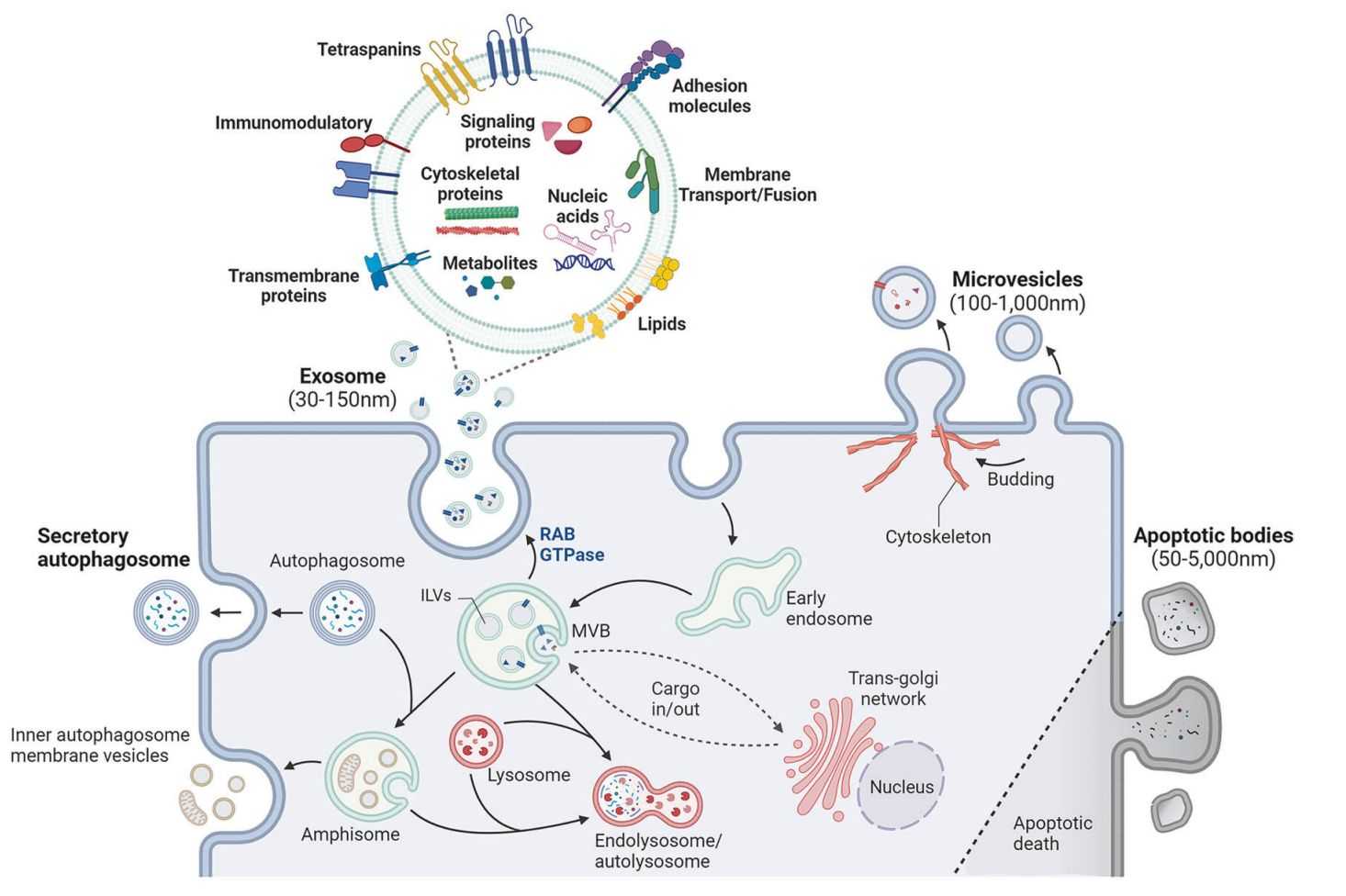
Exosomes carry a rich repertoire of functional proteins that regulate intercellular communication, immune modulation, tumor progression, and neurodegenerative pathways. As exosome research advances toward clinical translation, comprehensive and accurate proteomic profiling has become essential for understanding disease mechanisms, discovering biomarkers, and identifying therapeutic targets.
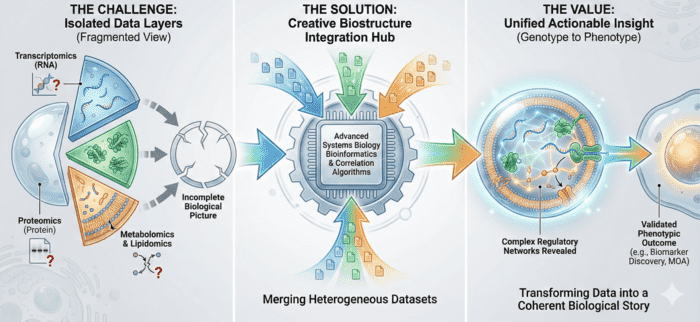
At Creative Biostructure, we provide an end-to-end Exosome Proteomics Services Platform integrating high-purity EV isolation, state-of-the-art mass spectrometry, targeted proteomic technologies, and publication-ready bioinformatics. Our workflows follow MISEV2023 recommendations and incorporate strict QC checkpoints to ensure confidence in exosome identity, purity, and analytical reproducibility.
Why Exosome Proteomics Matters
Exosomal proteins play essential roles in:
- Tumor microenvironment remodeling and metastasis
- Immune activation, suppression, and antigen presentation
- Neuronal signaling and neurodegenerative disease mechanisms
- Angiogenesis, extracellular matrix remodeling, and tissue repair
- Drug response, resistance mechanisms, and pathway reprogramming
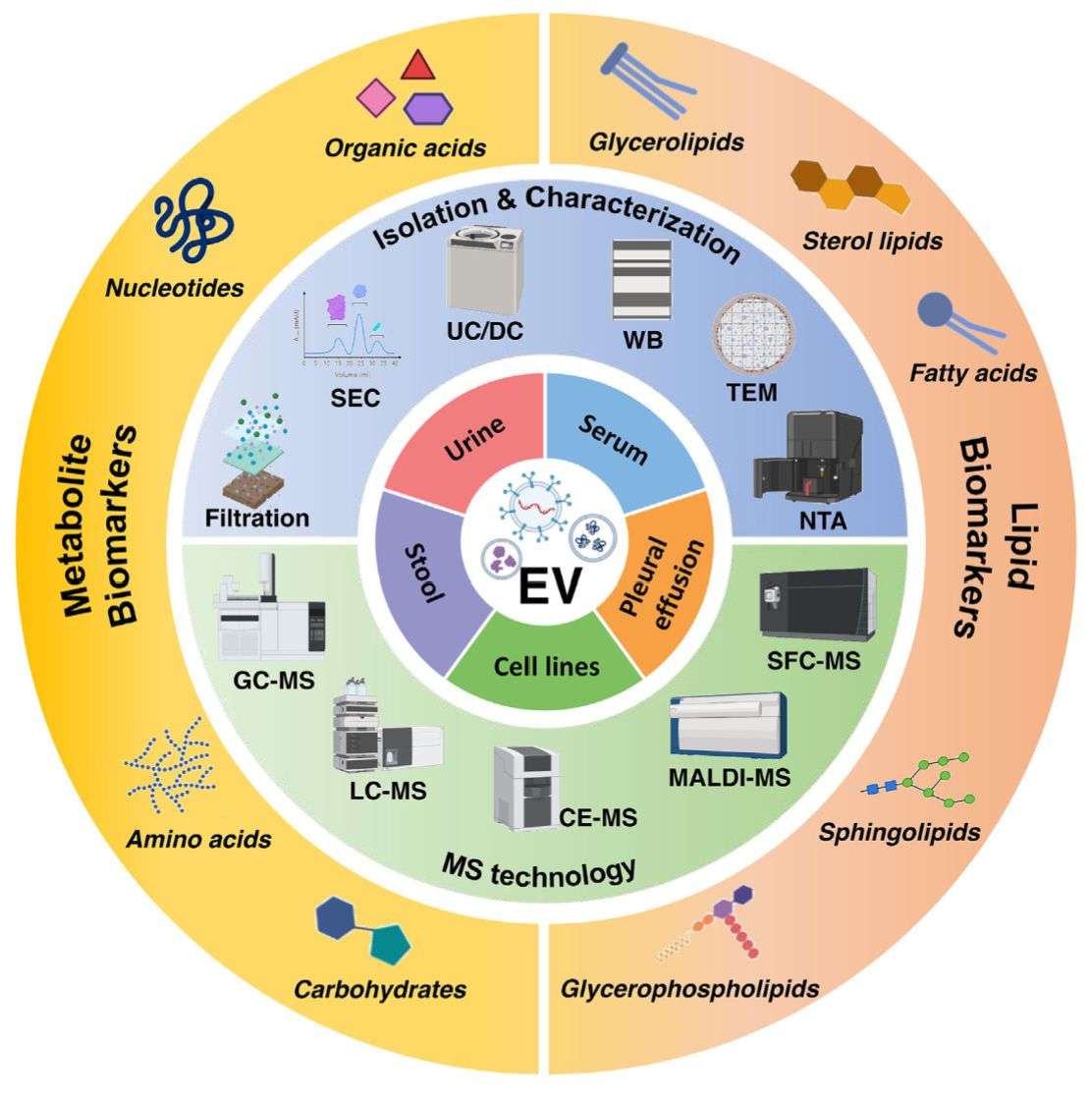
Because exosomes circulate in accessible fluids (plasma, serum, saliva, urine, CSF), proteomics allows non-invasive biomarker discovery and deep pathway exploration directly linked to disease biology.
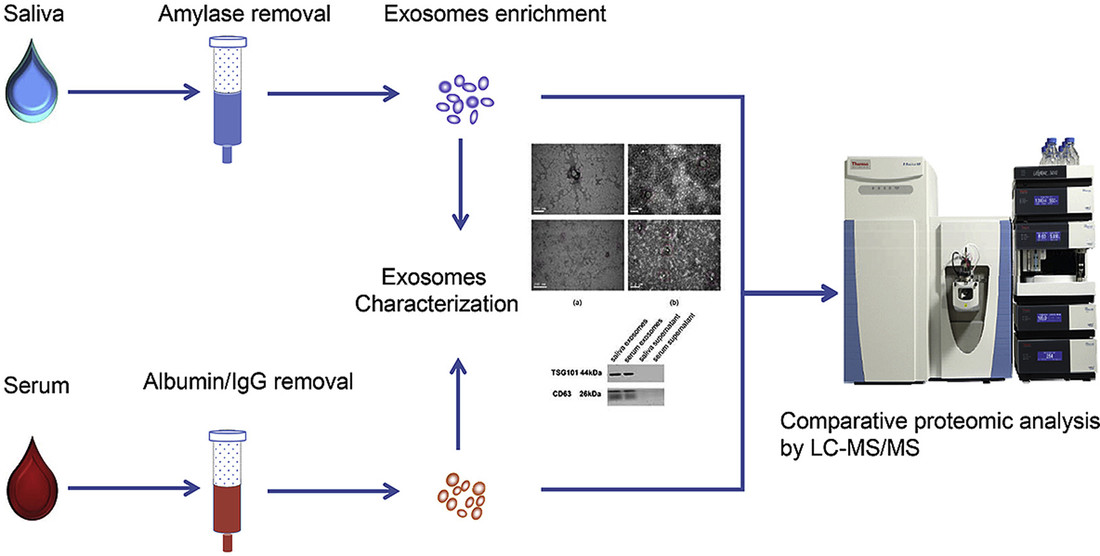 Figure 1. Systematic Comparison of Saliva- and Serum-Derived Exosomal Proteomes for Lung Cancer Detection. (Sun Y, et al., 2017)
Figure 1. Systematic Comparison of Saliva- and Serum-Derived Exosomal Proteomes for Lung Cancer Detection. (Sun Y, et al., 2017)
Our Exosome Proteomics Portfolio
Creative Biostructure provides a modular, scalable suite of specialized proteomic services:
Exosome Quantitative Proteomics
High-accuracy DDA/DIA or TMT-based quantification to characterize global expression changes across conditions, treatments, or disease states.
Exosome Biomarker Protein Screening
Targeted discovery of protein signatures associated with disease progression, treatment response, or patient stratification.
Single-Vesicle Membrane Proteomics
Ultra-sensitive profiling of membrane-associated proteins from individual vesicles to resolve EV heterogeneity and identify rare functional subsets.
Exosome Protein Profiling via Olink Technology
High-throughput antibody-based proximity extension assays (PEA) covering immune markers, inflammation signatures, neurology panels, and oncology pathways.
Integrated Exosome-Single Cell-Tissue Proteomics
Multi-omics analysis combining exosome proteome data with transcriptomic and proteomic maps from single-cell or spatial tissue profiling.
Exosomal Phosphoproteomics Analysis
Quantitative mapping of phosphorylation sites to investigate regulatory signaling networks transmitted via exosomes.
Exosomal Glycosylation Analysis
Structural characterization and quantification of glycan modifications on exosomal proteins to understand functional PTM regulation.
Our Workflow from Sample to Biological Interpretation
To ensure scientific integrity and reproducibility, every project follows a controlled, transparent workflow.
Sample Assessment and Pre Processing
We review sample type, volume, and matrix, remove major interfering components, and apply unified handling procedures to protect vesicle integrity before isolation.
High Purity Exosome Isolation
We choose suitable exosome isolation methods such as ultracentrifugation, gradient separation, size exclusion chromatography, ultrafiltration, or immunoaffinity and confirm purity by TEM, NTA, and exosome surface marker analysis.
Protein Extraction and Digestion
We lyse exosomes with protocols tuned for global and modified proteins, perform enzymatic digestion, and apply enrichment for phosphorylated or glycosylated peptides when required.
Mass Spectrometry and Targeted Proteomics
We analyze peptides on Orbitrap Fusion Lumos, Q Exactive HF X, timsTOF Pro 2 or Olink panels using DDA, DIA, TMT multiplexing, or targeted PEA to obtain sensitive and precise quantification.
Bioinformatics and Functional Annotation
We identify and quantify proteins, assess differential expression, perform PCA and clustering, and link results to GO, KEGG, Reactome, and exosome databases to reveal biological mechanisms.
Deliverables
You receive raw data, identification and quantitation tables, PTM information when relevant, quality control records, and an expert report with key findings and figures ready for publication.
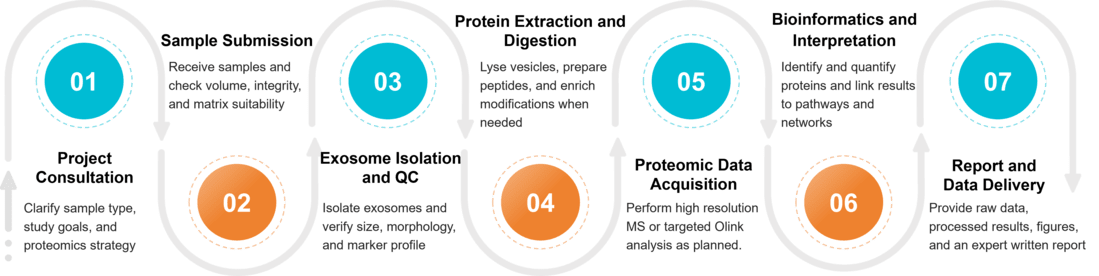 Figure 2. Project Workflow for Exosome Proteomics. (Creative Biostructure)
Figure 2. Project Workflow for Exosome Proteomics. (Creative Biostructure)
Sample Requirements
| Sample Type | Recommended Amount | Notes |
|---|---|---|
| Plasma/Serum | 3-5 mL | Avoid hemolysis; EDTA recommended |
| Urine | 10-20 mL | Early-morning preferred |
| Saliva | 2-5 mL | Remove amylase if biomarker-sensitive |
| Cell Culture Supernatant | 10-50 mL | Serum-free or EV-depleted media |
| FFPE | 10-20 slides | 10 μm thickness |
| Tissue | 50-200 mg | Fresh or snap-frozen |
Reporting & Deliverables (Publication-Ready)
You receive a complete, publication-ready data package:
- Raw MS data files
- Peptide/protein identification tables
- Quantitative expression matrices
- PTM site information (if applicable)
- Full quality control documentation
- Interpretive report with biological insights
- High-resolution figures for manuscripts and grant proposals
Key Research Areas Supported by Exosome Proteomics
- Cancer Biology: Exosome proteomics helps identify early cancer indicators, clarify metastatic pathways, study interactions between tumor and surrounding tissues, and evaluate treatment response using circulating protein signatures.
- Immunology: Analysis of exosomal proteins reveals cytokine and chemokine cargo, protein factors involved in antigen presentation, and immune activation patterns that shape inflammatory and regulatory processes.
- Neurology and Neurodegeneration: Protein profiling of neural derived exosomes highlights synaptic and lysosomal changes, detects markers of central nervous system inflammation, and supports early study of Alzheimer's and Parkinson's diseases.
- Cardiovascular and Metabolic Diseases: Exosomes carry proteins linked to cardiac stress and remodeling, as well as enzymes involved in metabolic regulation. Their profiling helps track disease progression and metabolic dysfunction.
- Drug Discovery and Mechanistic Research: Exosome proteomics uncovers protein changes linked to therapy resistance, identifies treatment influenced pathways, and supports evaluation of drug effects on cell signaling networks.
Why Choose Creative Biostructure
- MISEV2023-Aligned EV Characterization: Ensures the highest standard of EV purity and identity.
- Deep Proteome Coverage: Optimized extraction and MS platforms support detection of thousands of proteins, including low-abundance markers.
- Multi-Modal Analytical Options: Combine MS-based proteomics with Olink, PTM analysis, or single-vesicle profiling.
- Expert Bioinformatics Interpretation: Data is not just delivered; it is interpreted through biological context by senior proteomics scientists.
- Tailored Protocols for Complex Samples: Extensive experience with plasma, CSF, urine, saliva, supernatants, FFPE tissue, and plant/microbial EVs.
Case Study
Case: CSF Exosomal Proteomics Identifies C1q as an Early AD Marker
Background
CSF extracellular vesicles from Alzheimer's disease patients and non-neurodegenerative controls were profiled to discover EV protein biomarkers linked to early neuroinflammation.
Methods
- EV Identification: CSF EVs were isolated by ultracentrifugation or Vn96 affinity capture. Vesicle identity and purity were confirmed through TEM, NTA, and EV marker assays following MISEV guidelines.
- Proteome Profiling: Untargeted label-free LC-MS (DIA or DDA depending on cohort) quantified CSF EV proteins across two independent discovery cohorts.
- Validation: ELISA was performed in two additional cohorts to validate top candidates, particularly complement protein C1q.
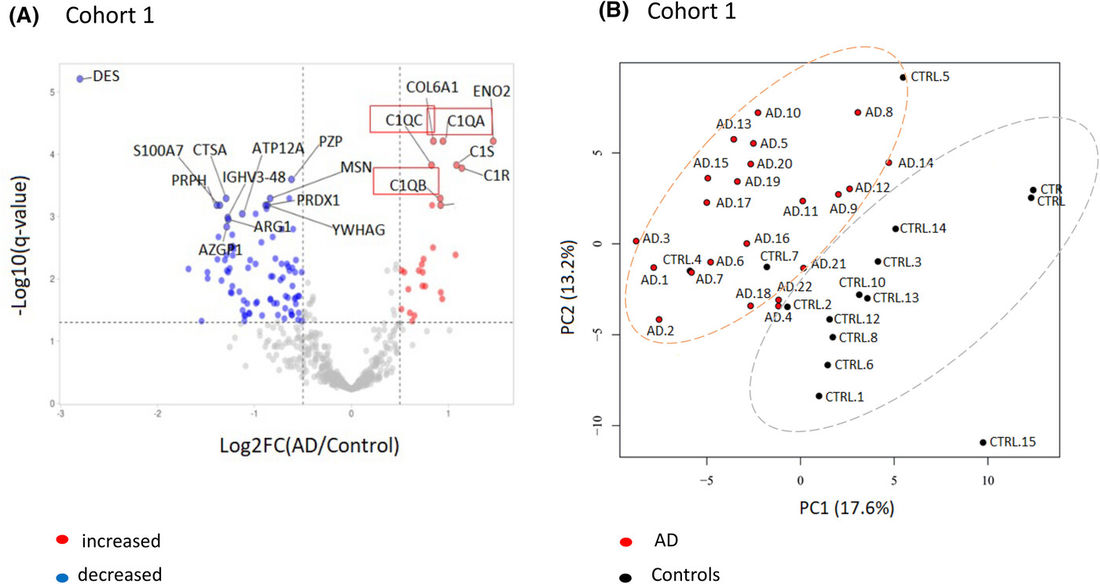 Figure 3. Proteomic Differences in Alzheimer's Disease (AD). (A) Volcano plot comparing AD vs. non-demented controls in Cohort 1; C1q is increased (red boxes). Thresholds: log2FC ≥ 0.5 and −log10(p) ≥ 1.3. (B) PCA shows clear separation between AD (red) and controls (black), indicating distinct proteomic profiles. (Chatterjee M, et al., 2023)
Figure 3. Proteomic Differences in Alzheimer's Disease (AD). (A) Volcano plot comparing AD vs. non-demented controls in Cohort 1; C1q is increased (red boxes). Thresholds: log2FC ≥ 0.5 and −log10(p) ≥ 1.3. (B) PCA shows clear separation between AD (red) and controls (black), indicating distinct proteomic profiles. (Chatterjee M, et al., 2023)
Results
More than 30 EV proteins were differentially expressed in AD, with complement components, especially C1q, consistently increased. ELISA confirmed higher EV C1q in AD and MCI due to AD versus controls, with significant group separation.
Conclusion
The study shows that CSF EV associated C1q is a robust early inflammatory marker in Alzheimer's disease. It illustrates how exosome proteomics plus orthogonal validation can yield clinically relevant CNS biomarkers beyond bulk CSF analysis.
Ready to translate exosomal protein data into clear biological insight? Whether your objective is deep proteome discovery, quantitative differential analysis, post-translational modification (PTM) profiling, or protein biomarker screening, our specialized proteomics solutions deliver high-resolution insights tailored to your study design. Contact us for a brief consultation and a fast, customized quotation.
References
- Sun Y, Liu S, Qiao Z, et al. Systematic comparison of exosomal proteomes from human saliva and serum for the detection of lung cancer. Analytica Chimica Acta. 2017, 982: 84-95.
- Chatterjee M, Özdemir S, Kunadt M, et al. C1q is increased in cerebrospinal fluid‐derived extracellular vesicles in Alzheimer's disease: A multi‐cohort proteomics and immuno‐assay validation study. Alzheimer's & Dementia. 2023, 19(11): 4828-4840.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.