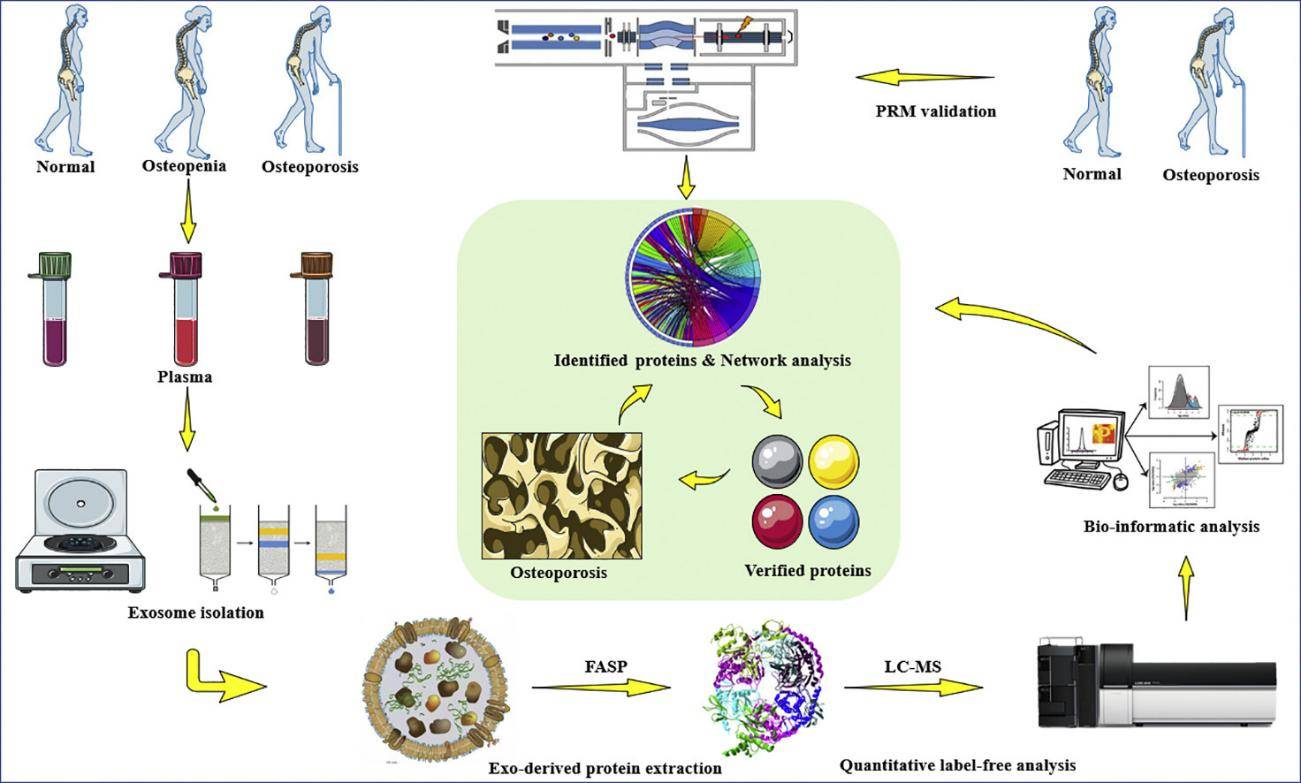
Single-Vesicle Membrane Proteomics Services
Single-vesicle membrane proteomics service leverages cutting-edge technologies to conduct high-resolution, precise profiling of the protein landscape on the surface of individual extracellular vesicles. This groundbreaking approach serves as a pivotal tool for uncovering novel surface biomarkers, understanding intercellular communication mechanisms, and advancing targeted therapeutic development. Creative Biostructure provides state-of-the-art single-vesicle analysis that deciphers the functional membrane proteome with unparalleled detail, delivering critical insights for fundamental biomedical research and theranostic applications.
What are The Advantages of Single-Vesicle Membrane Proteomics?
Vesicles serve as core carriers for intracellular material transport and signal transduction. The composition and function of their membrane proteins, which directly determine cellular physiological and pathological states, are critical in diverse processes. These range from synaptic neurotransmission to the distal regulation of tumors mediated by exosomes. Therefore, deciphering the heterogeneity of vesicle membrane proteins is key to understanding the microscopic mechanisms of life.
Traditional bulk vesicle proteomics fails to capture membrane protein differences between individual vesicles due to averaging effects. In contrast, single-vesicle membrane proteomics services integrate a comprehensive technical system encompassing "single-vesicle precision separation - efficient membrane protein extraction - high-sensitivity mass spectrometry detection - multidimensional bioinformatics analysis," has pioneered the qualitative, quantitative, and functional analysis of membrane proteomes in individual vesicles. This revolutionary tool unlocks insights into vesicle heterogeneity mechanisms and facilitates the discovery of novel biomarkers and drug targets.
Our Single-Vesicle Membrane Proteomics Services
Creative Biostructure provides clients with a one-stop single-vesicle membrane proteomics solution, from sample preparation to bioinformatics analysis. Our services include but are not limited to the following:
| Services | Description |
|---|---|
| Single-Vesicle Isolation and Enrichment | Tailor-made separation strategies for vesicles of different origins and sizes:
|
| Single-Vesicle Membrane Protein Extraction and Purification | To address the characteristics of membrane proteins, we employ an optimized extraction system:
|
| High-Sensitivity Mass Spectrometry Detection |
|
| Proteomics Bioinformatics Analysis | We deliver multi-dimensional data analysis customized to our clients' specific needs:
|
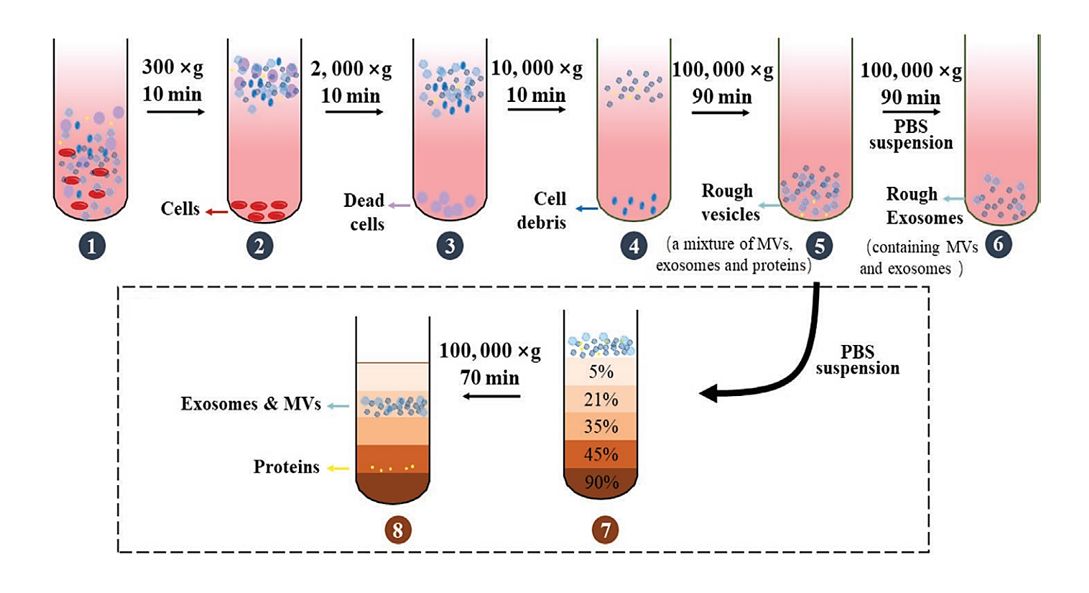
Workflow of Single-Vesicle Membrane Proteomics Analysis
We adhere to standardized and traceable experimental protocols to ensure reliable results at every step.
Sample Receipt and Evaluation
Accepted Sample Types: Fresh/frozen vesicle suspensions, cell lysates (vesicles must be pre-isolated by the client), bodily fluid samples (serum, cerebrospinal fluid, cell culture supernatants).
Sample Evaluation: Determine vesicle concentration (NTA nanoparticle tracking analysis), purity (Western Blot for contaminating proteins), and confirm experimental objectives with the client (e.g., qualitative/quantitative analysis, differential analysis direction).
Single-Vesicle Preparation and Quality Control
Select separation methods (microfluidics/magnetic bead capture) based on sample characteristics to isolate individual vesicles.
Verify single-vesicle integrity via AFM/CLSM and issue quality control reports.
Membrane Protein Extraction and Mass Spectrometry Analysis
Optimize the detergent system for membrane protein extraction, followed by concentration and nano-LC-MS/MS analysis.
Continuously monitor mass spectrometry data quality (e.g., peptide matching rate, protein coverage) to ensure data integrity.
Bioinformatics Analysis and Report Delivery
We perform multidimensional data analysis based on client requirements and generate visualization charts (e.g., volcano plots, heatmaps, pathway enrichment plots).
A comprehensive report is delivered, including: the experimental protocol, quality control data, raw mass spectrometry files (.raw), analysis result tables, figures, and biological interpretation of conclusions.
 Figure 1. Single-Vesicle Membrane Proteomics Analysis Workflow. (Creative Biostructure)
Figure 1. Single-Vesicle Membrane Proteomics Analysis Workflow. (Creative Biostructure)
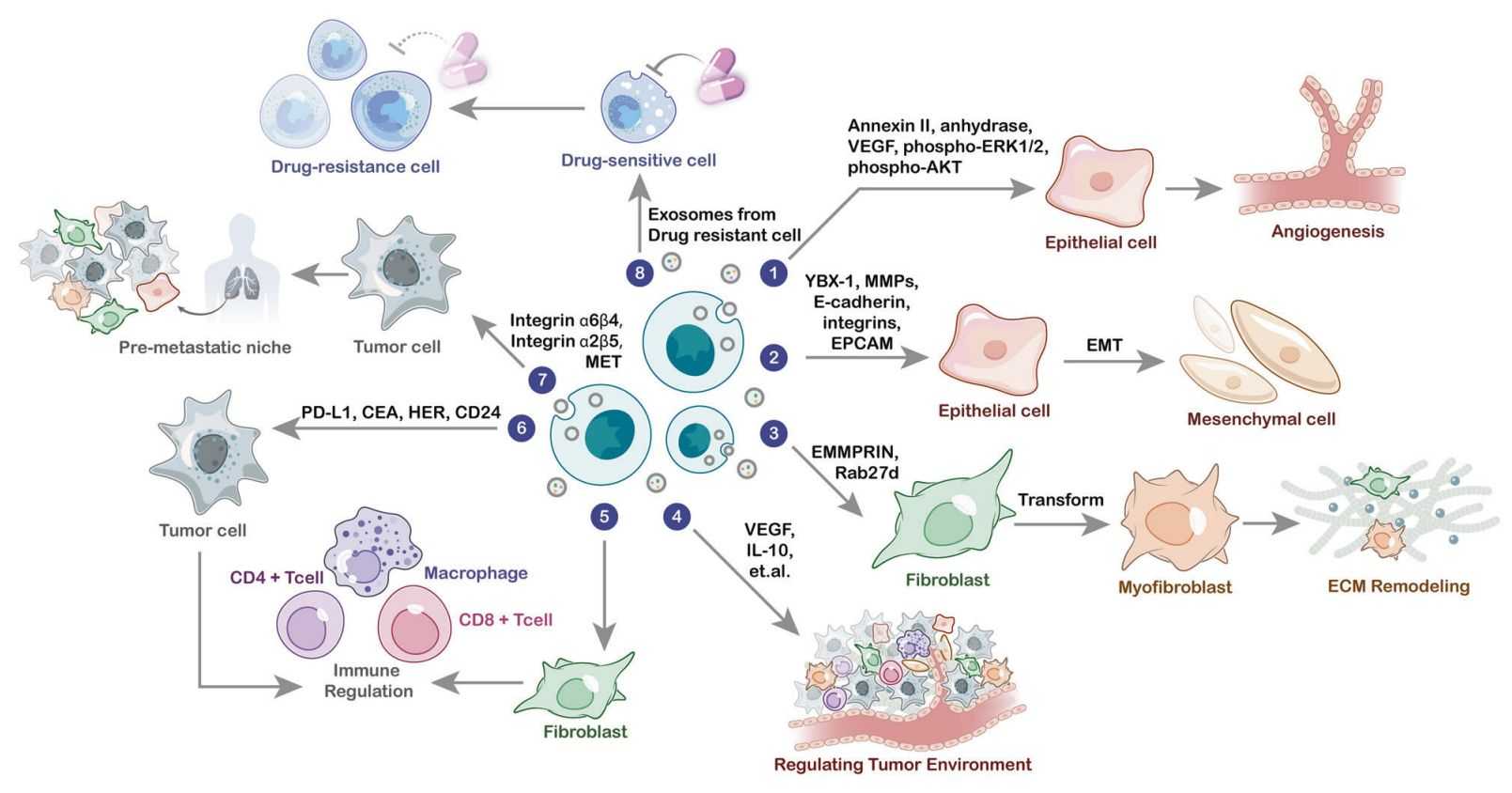
Application of Single-Vesicle Membrane Proteomics Analysis
- Neuroscience
Elucidating the relationship between synaptic vesicle membrane protein heterogeneity and neurotransmitter release.
Investigating the sorting mechanism of amyloid precursor protein (APP) into single vesicles in Alzheimer's disease (AD).
- Tumor Biology
Screening for tumor-derived exosomal single-vesicle membrane protein biomarkers (e.g., PD-L1, CD47) for early diagnosis.
Analyzing changes in single-vesicle membrane proteins of tumor cell exosomes after chemotherapy to predict drug resistance mechanisms.
- Cell Biology
Investigating the sorting and trafficking of vesicle membrane proteins in the endolysosomal pathway.
Elucidating the impact of cellular stress (e.g., hypoxia, oxidative stress) on the composition of single-vesicle membrane proteins.
- Drug Discovery
Identifying drug targets on single-vesicle membrane proteins (e.g., ion channels, transporters).
Evaluating the regulatory effects of drugs on single-vesicle membrane protein expression to optimize dosing regimens.
- Cardiovascular Diseases
Analyzing the role of endothelial cell vesicle single-vesicle membrane proteins (e.g., eNOS) in vascular homeostasis.
Screening for single-vesicle membrane protein biomarkers in serum exosomes of myocardial infarction patients for prognostic evaluation.
Customer Support & Delivery
- Client Requirements
Samples: Fresh/frozen vesicle samples (must specify sample type, processing method, and storage conditions);
Experimental Requirements: Clearly define analysis objectives (qualitative/quantitative, control group design, pathways or proteins of interest);
Special Requirements: Such as customized data analysis content, report formats, etc.
- Deliverables
Raw experimental data: Mass spectrometry .raw files, SDS-PAGE/Western Blot raw images, AFM/CLSM quality control images.
Analysis report: Includes experimental methods, quality control results, protein identification/quantification lists, bioinformatics analysis charts and conclusions.
Supplementary materials: Technical methodology descriptions, software parameters used for data analysis.
Case Study
Case: Precise Trafficking of a Synaptic Vesicle Protein Subset Revealed by Single-Vesicle Proteomics.
Background
Protein sorting regulates neurotransmission by defining synaptic vesicle composition. While bulk studies have estimated average protein numbers, intervesicle variability and sorting precision remain unknown. To address this, we employed a single-molecule approach to quantify the abundance and variance of seven integral membrane proteins in brain synaptic vesicles.
Methods
- Isolation of vesicles from brain homogenates
- Preparation of vesicles for fluorescence imaging
- Fabrication of micro-channels
- Optical setup
- Fluorescent imaging of single vesicles and single antibodies
- Fluorescent puncta analysis
- Data analysis and fitting
Conclusion
This study reports that four vesicular proteins (SV2, the proton ATPase, Vglut1, and synaptophysin 1) exhibit minimal intervesicular variation in quantity, indicating their high-precision sorting into vesicles. In contrast, the apparent numbers of VAMP2/synaptobrevin 2, synaptotagmin, and synapsin demonstrated substantial variability between vesicles. These findings impose constraints on protein-functional models of the synapse and suggest that differences in vesicular protein expression may influence vesicle composition and function.
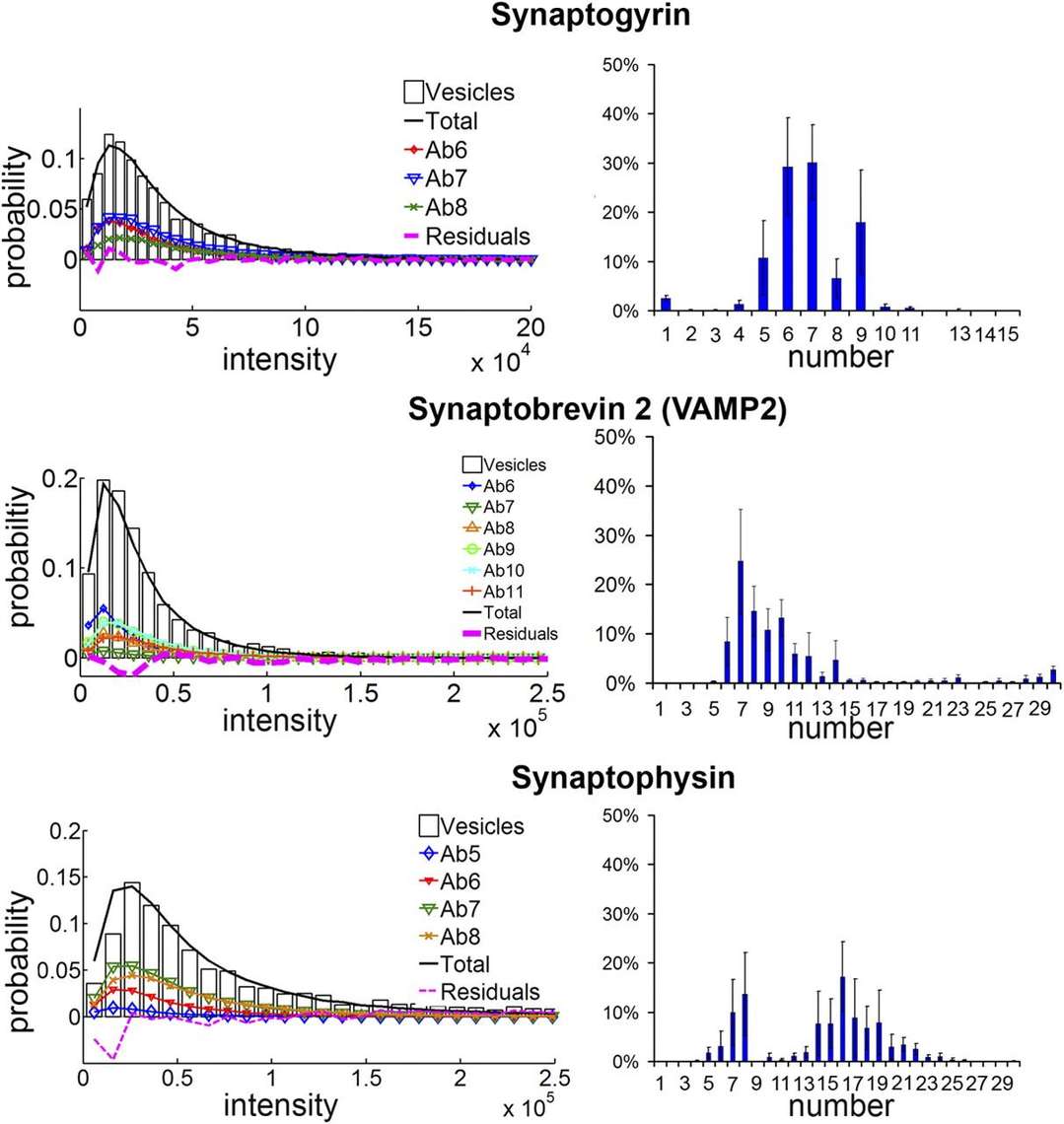 Figure 2. Quantification of polydispersed synaptic vesicle proteins. (Mutch S A, et al., 2011)
Figure 2. Quantification of polydispersed synaptic vesicle proteins. (Mutch S A, et al., 2011)
Ready to resolve vesicle heterogeneity one membrane at a time? Our single-vesicle membrane proteomics workflow combines precision isolation, high-sensitivity MS, and multidimensional bioinformatics to reveal actionable surface biomarkers and targets. Contact us to tailor the pipeline to your samples and get a fast, study-ready quotation.
Reference
- Mutch S A, Kensel-Hammes P, Gadd J C, et al. Protein quantification at the single vesicle level reveals that a subset of synaptic vesicle proteins are trafficked with high precision. Journal of Neuroscience. 2011, 31(4): 1461-1470.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.