Exosome Lipidomics Services
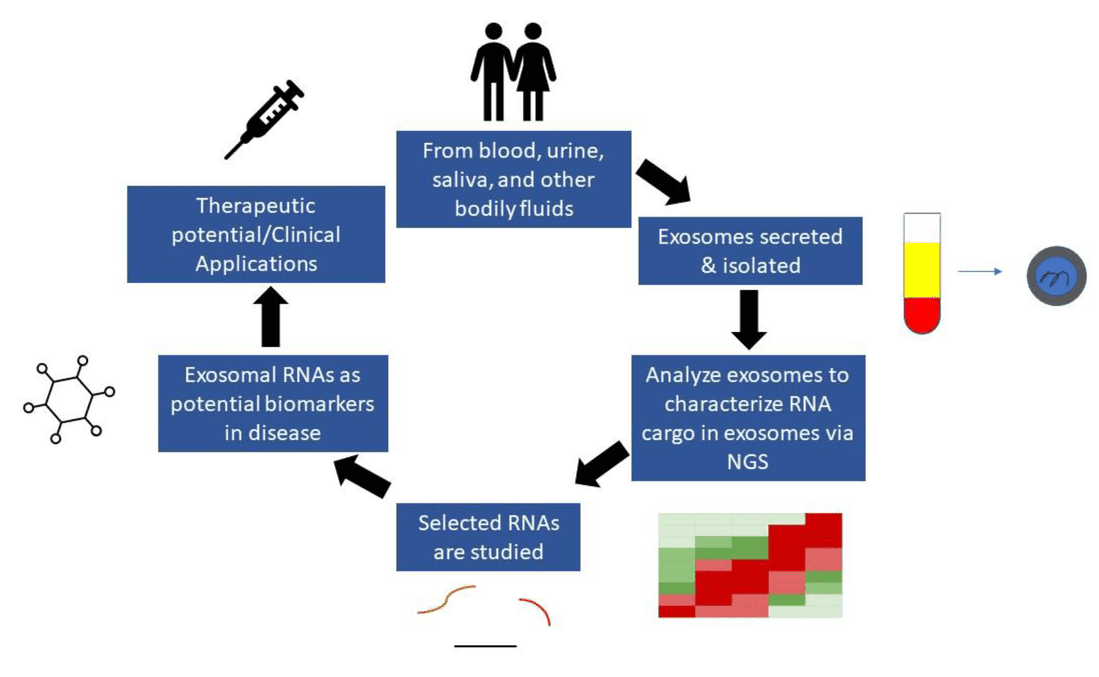
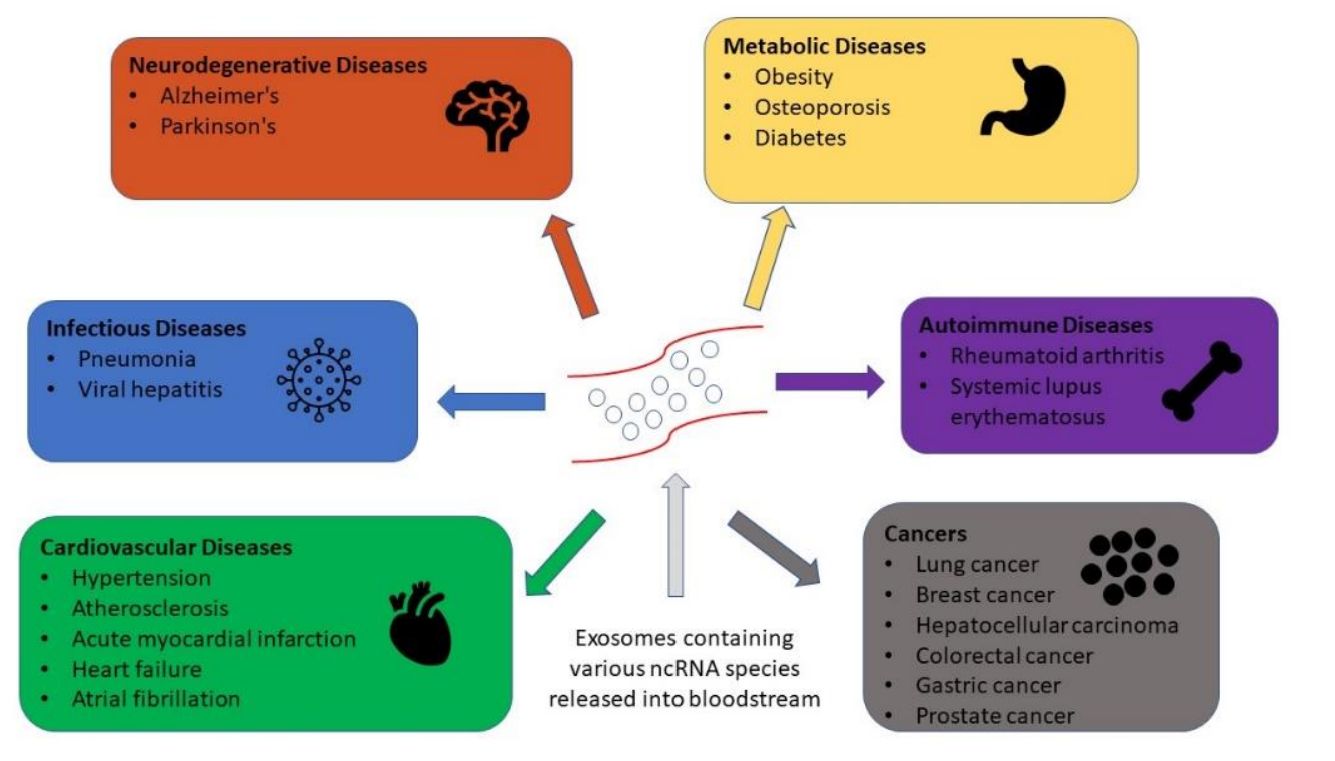
Exosomes carry a rich repertoire of bioactive molecules, including proteins, nucleic acids, metabolites, and lipids, that reflect the physiological state of their parent cells. Among these components, lipids play a fundamental role in vesicle formation, membrane stability, intercellular communication, and disease signaling. At Creative Biostructure, we offer comprehensive exosome lipidomics services designed to decode lipid composition and function across diverse biological sources.
What Is Exosome Lipidomics?
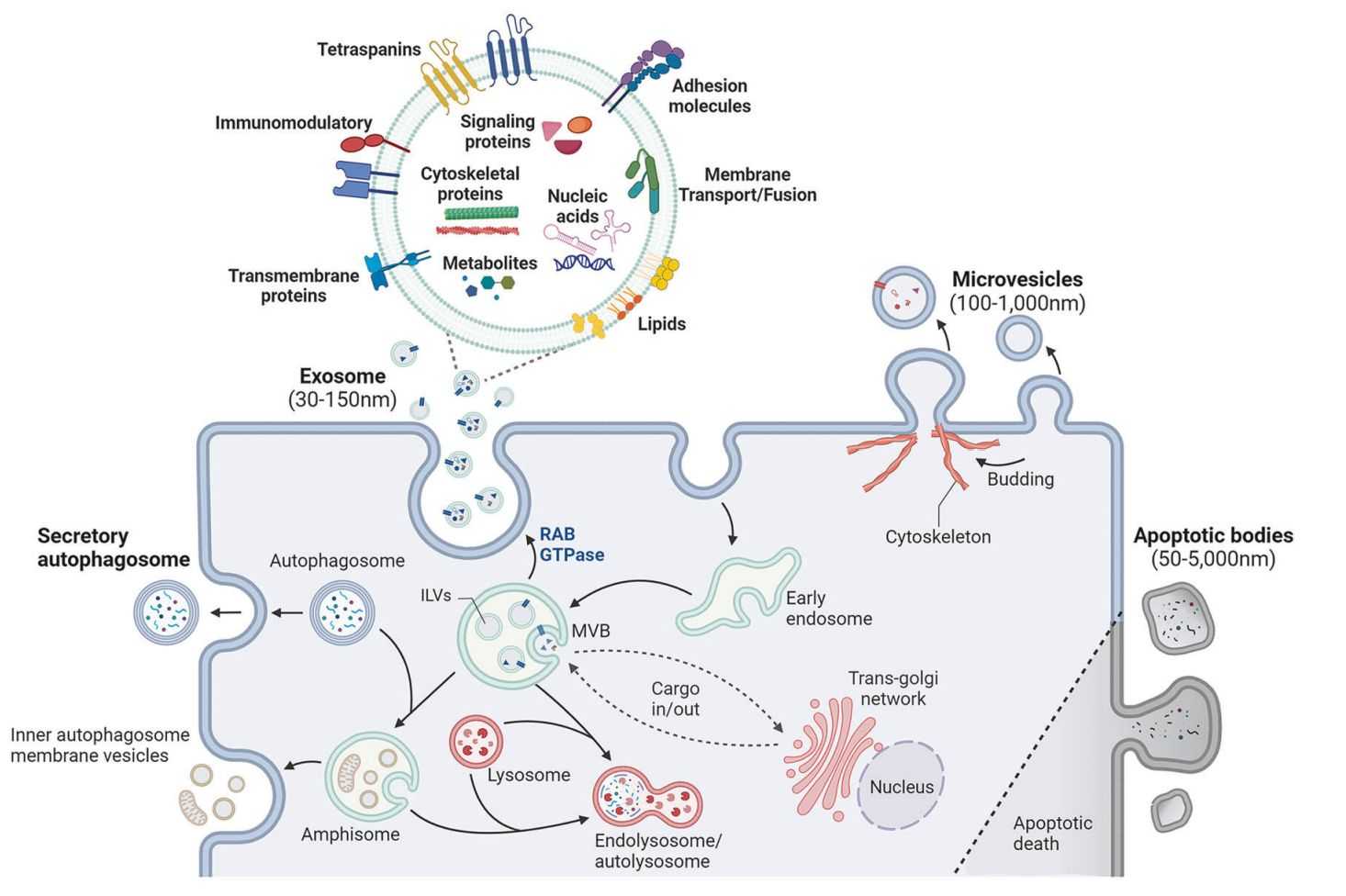
Exosome lipidomics is the systematic profiling of lipid species contained within exosomes, small extracellular vesicles typically 30-150 nm in diameter. These vesicles originate from multivesicular body (MVB) maturation and are secreted into the extracellular space, where they act as carriers of molecular messages.
Exosomal lipids contribute to:
- Membrane structure and rigidity (e.g., enriched sphingolipids and cholesterol)
- Vesicle biogenesis, including budding and cargo sorting
- Ligand-receptor interactions with target cells
- Activation of intracellular signaling pathways
- Disease-associated lipid remodeling, which provides biomarkers for diagnostics and monitoring
Understanding exosomal lipid composition provides valuable clues to cellular function, immune activity, stress responses, and pathological conditions.
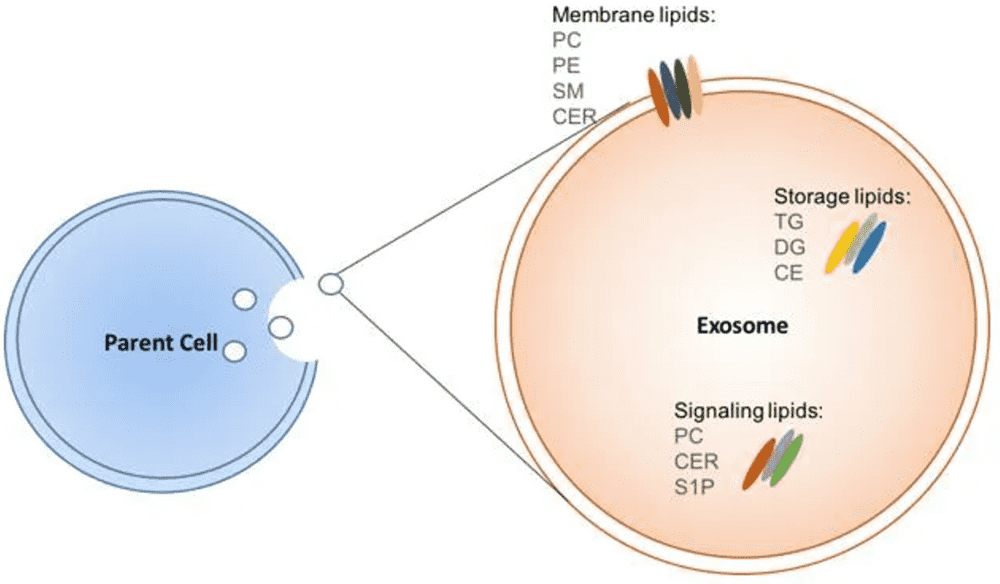 Figure 1. Schematic of Exosome Lipid Composition. (Javdani-Mallak A, et al., 2024)
Figure 1. Schematic of Exosome Lipid Composition. (Javdani-Mallak A, et al., 2024)
What Our Exosome Lipidomics Services Deliver
Our service is designed to generate reproducible, high-accuracy lipidomic profiles with deep biological interpretation.
Lipid Classes We Analyze
Below is a non-exhaustive list of lipid categories included in our profiling:
| Lipid Class | Representative Lipid Species |
|---|---|
| Phospholipids |
|
| Sphingolipids |
|
| Glycerolipids |
|
| Sterols |
|
| Fatty Acids |
|
| Lysophospholipids |
|
| Other Categories |
|
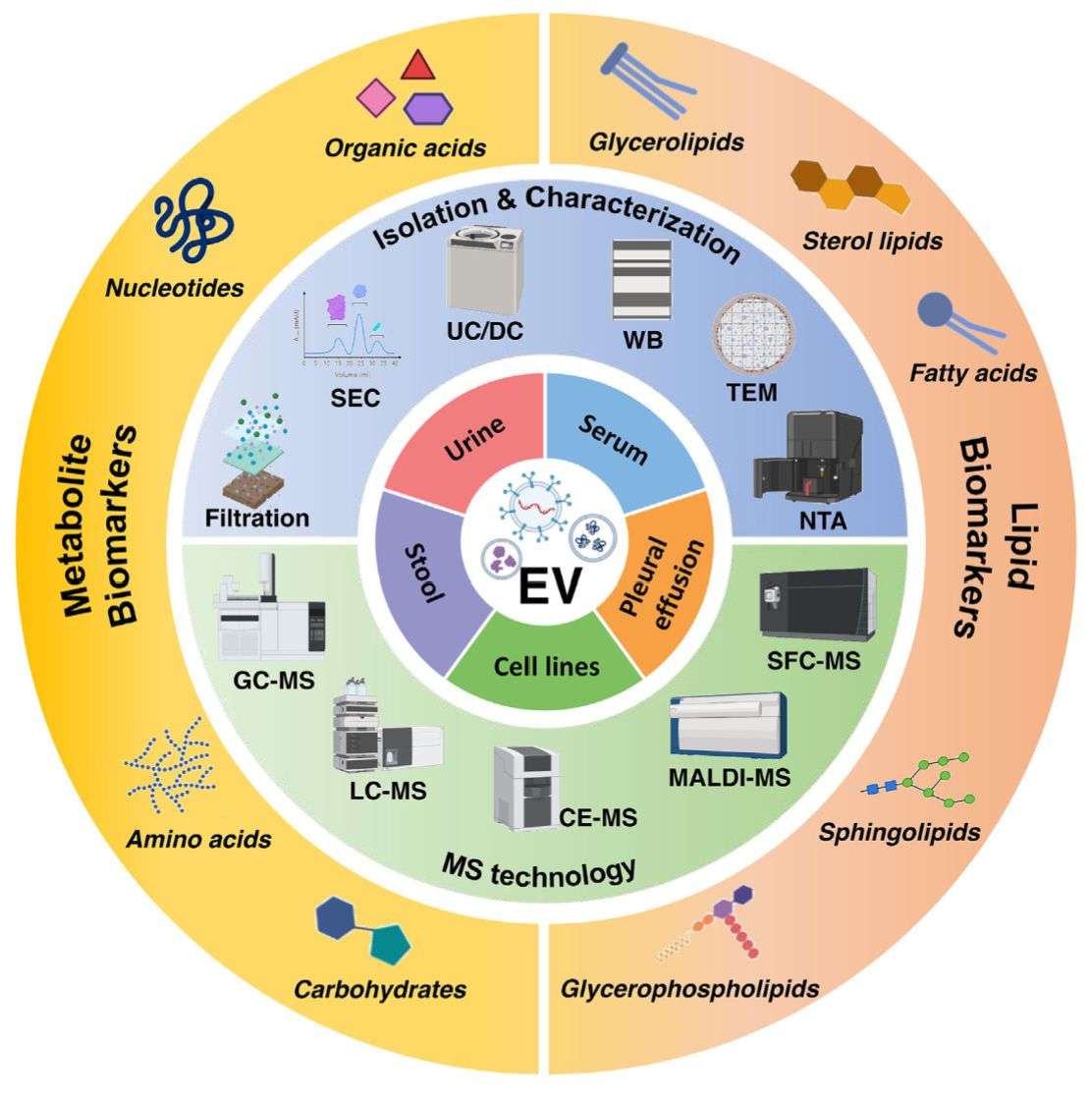
Technology Platforms
To ensure high-depth, high-accuracy exosome lipid profiling, Creative Biostructure integrates multiple complementary mass spectrometry (MS) platforms.
| LC-MS / LC-MS/MS (Orbitrap & Q-TOF) Ultra-high resolution identification and quantification of complex lipid species. |
UHPLC Superior chromatographic separation for isobaric and isomeric lipids. |
| Shotgun Lipidomics (Direct Infusion-MS) High-throughput lipid class coverage for large sample cohorts. |
GC-MS Accurate quantification of fatty acids and volatile lipid species. |
Instrument examples may include:
- Orbitrap Exploris or Q Exactive series
- TripleTOF/Q-TOF systems
- High-end UHPLC units (Agilent / Thermo / Waters)
- Agilent GC-MS systems
(Note: Models listed are representative of technologies used and may vary by project.)
Explore Our Source-Specific Exosome Lipidomics Services
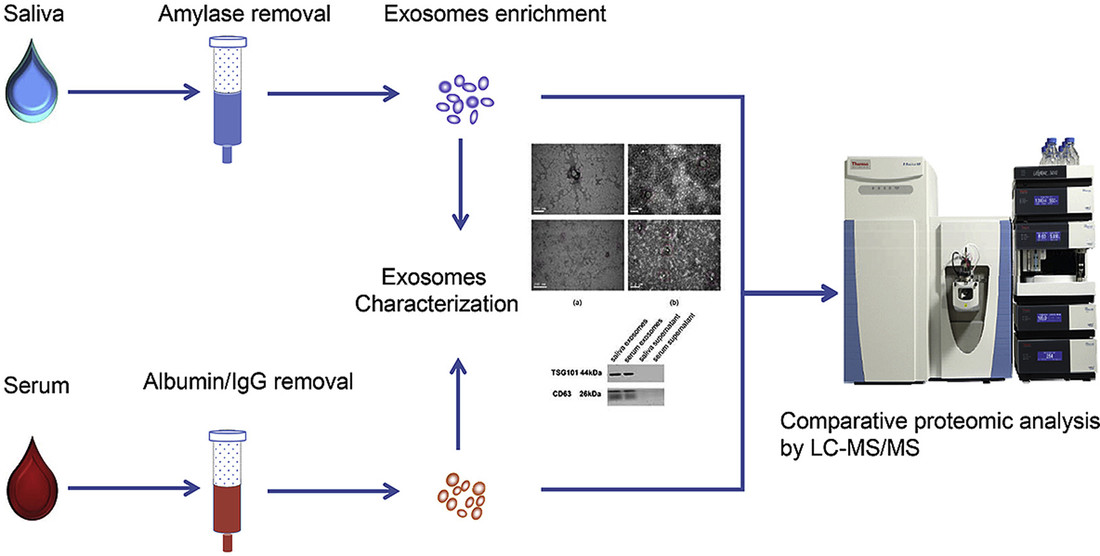
Creative Biostructure offers a series of specialized, sample-type-specific exosome lipidomics workflows. Each service integrates optimized exosome isolation strategies, tailored lipid extraction chemistries, and advanced LC-MS/MS platforms to ensure the most accurate and biologically meaningful lipid data.
Human exosome lipidomics from diverse biofluids supports biomarker discovery, disease and immune profiling, and drug response studies, with sensitive detection and optional multi-omics.
Rodent exosome lipidomics links disease progression, therapy effects, and metabolism in preclinical models, supporting toxicity, efficacy, and dose-response studies.
Milk exosome lipidomics profiles bioactive lipids for nutrition, immunity, infant development and oral EV delivery, supporting food, dairy and agricultural research.
Plant exosome-like nanoparticle lipidomics reveals unique plant lipids for natural product, anti-inflammatory, drug delivery and agricultural resilience research.
Yeast exosome lipidomics maps lipids driving fungal signaling, stress adaptation, pathogenicity and fermentation, supporting bioprocess optimization and fungal research.
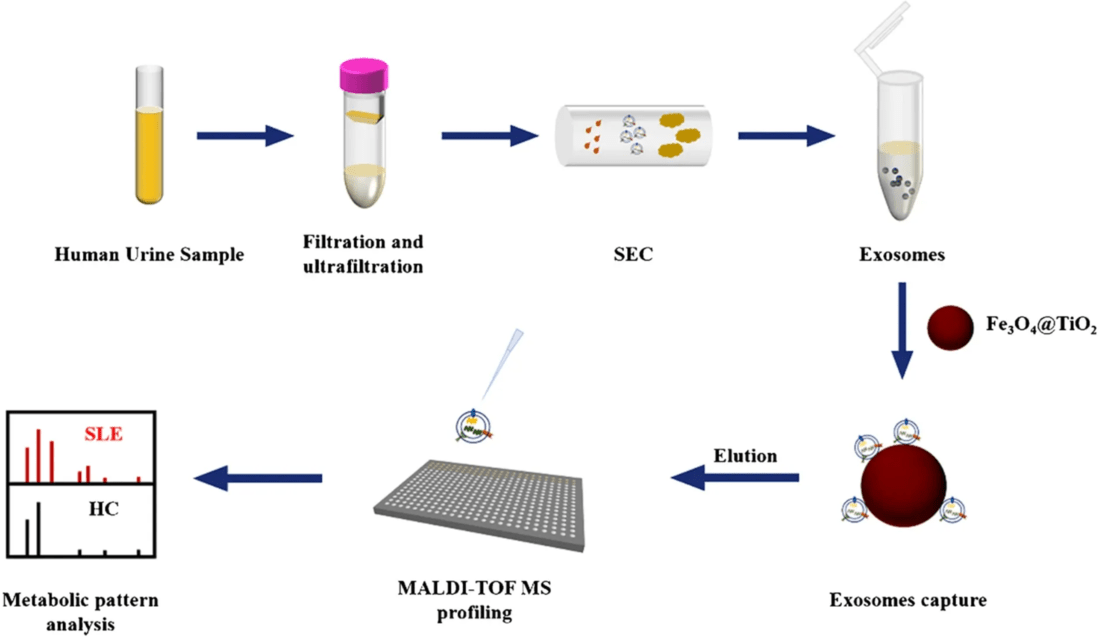
Exosome Lipidomics Workflow
Our workflow has been designed for accuracy, reproducibility, and transparency.
Project Consultation & Study Design
We clarify research goals, sample types, and analytical depth to build a tailored lipidomics plan that ensures accuracy and biological relevance.
Exosome Isolation & Purification
Exosomes are isolated using optimized UC, SEC, TFF, or immunocapture workflows to ensure high purity and consistency across all sample types.
Vesicle Quality Assessment
Particle size, morphology, protein markers, and purity are evaluated to confirm exosome integrity and suitability for downstream lipid analysis.
Lipid Extraction & MS-Based Profiling
Lipids are extracted with optimized chemistries and profiled using LC-MS, GC-MS, or shotgun MS for sensitive, high-coverage lipid identification.
Data Analysis, Interpretation & Reporting
Normalized datasets, statistics, and pathway insights are compiled into a clear, publication-ready report with optional expert consultation.
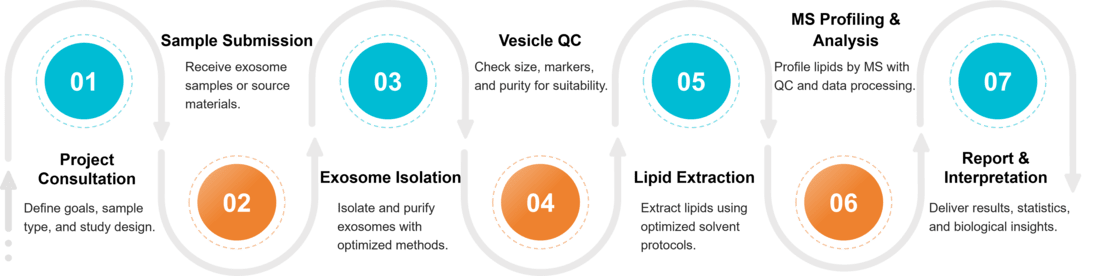 Figure 2. Project Workflow for Exosome Lipidomics. (Creative Biostructure)
Figure 2. Project Workflow for Exosome Lipidomics. (Creative Biostructure)
Sample Requirements
To ensure optimal lipid detection and quantitative reliability:
- Purity: Exosome samples must be free from debris, protein aggregates, and non-EV contaminants
- Storage: -80°C, minimal freeze-thaw cycles
- Volume / biomass varies by source (guidelines provided per sample type)
- Shipping: Dry ice transport recommended
Our team can provide custom exosome isolation solutions if needed.
Reporting & Deliverables (Publication-Ready)
You will receive a complete, publication-ready package including:
- Full list of identified lipid species
- Quantitative tables (absolute or relative)
- PCA / PLS-DA / heatmaps / volcano plots
- Lipid pathway enrichment
- Interpretation of biological significance
- Technical methodology and QC documentation
- Raw data files (optional)
Biological and Research Applications
Our exosome lipidomics solutions support a broad range of research objectives:
- Disease biomarker identification (cancer, neurodegeneration, cardiometabolic disorders)
- Mechanistic studies related to EV biogenesis and trafficking
- Therapeutic monitoring & drug response evaluation
- Host-pathogen interaction research
- Nutritional and agricultural studies (milk/plant exosomes)
- Industrial biotechnology (yeast-derived EVs)
Why Choose Creative Biostructure
- Comprehensive lipid class coverage: Phospholipids, sphingolipids, sterols, glycerolipids, fatty acids, lysophospholipids, ether lipids, gangliosides, and more
- High sensitivity & precise quantification: Enabled by Orbitrap and Q-TOF MS platforms
- Absolute & relative quantification: Using isotope-labeled internal standards
- Advanced bioinformatics: Including pathway mapping, network analysis, clustering, and PCA/PLS-DA
- Customizable workflows: Suitable for discovery research, translational studies, and therapeutic development
Case Study
Case: Cell-Type-Specific EV Lipidomics in Neurodegeneration
Background
Plasma EVs enriched for neuronal (L1CAM+), astrocytic (GLAST+), and pan-EV (CD9/CD63/CD81+) origins were profiled to explore lipid changes linked to brain aging and neurodegeneration.
Methods
Crude EVs were immunocaptured into three subsets, checked by NTA for size and concentration, then subjected to modified Bligh-Dyer extraction and targeted UPLC-MS/MS to quantify major phospholipid and sphingolipid classes.
Results
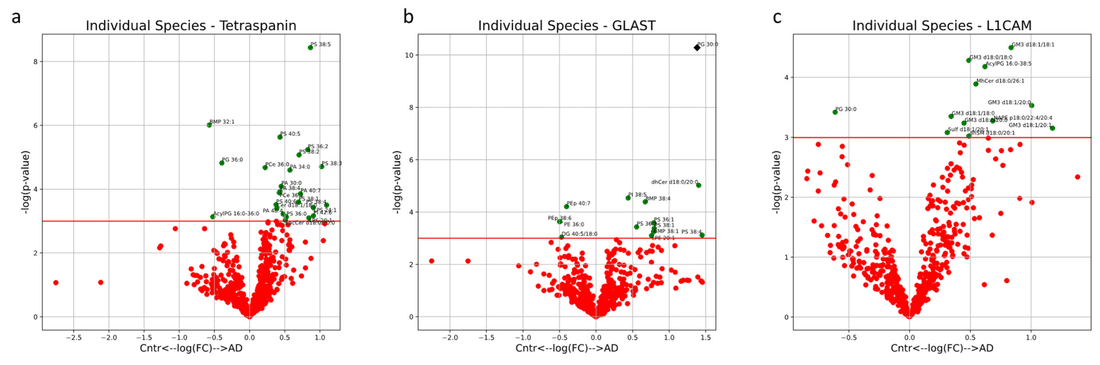 Figure 3. Volcano plots showing lipid concentration differences between AD and control EVs isolated by (a) pan-tetraspanin, (b) GLAST, and (c) L1CAM immunoprecipitation. Green points above the red line: p<0.05. Black diamonds: Bonferroni-significant (p<0.05). Positive log2 fold changes indicate enrichment in AD. (Krokidis M G, et al., 2024)
Figure 3. Volcano plots showing lipid concentration differences between AD and control EVs isolated by (a) pan-tetraspanin, (b) GLAST, and (c) L1CAM immunoprecipitation. Green points above the red line: p<0.05. Black diamonds: Bonferroni-significant (p<0.05). Positive log2 fold changes indicate enrichment in AD. (Krokidis M G, et al., 2024)
Neuronal- and astrocytic-enriched EVs showed distinct lipid patterns compared with pan-EVs, with specific phospholipids and sphingolipids altered in ways consistent with synaptic dysfunction, membrane remodeling, and neuroinflammatory pathways.
Conclusion
This case illustrates how combining cell-type-specific EV enrichment with targeted lipidomics can reveal subtle, CNS-relevant lipid alterations that are not detectable in bulk plasma, supporting EV lipidomics as a peripheral window into brain pathology.
Ready to turn exosomal lipid profiles into meaningful biological insight? Whether your goal is biomarker discovery, mechanistic pathway analysis, multi-source EV comparison, or high-precision lipid quantification, our advanced mass spectrometry platforms and expert analytical team deliver the depth and clarity your project demands. Contact us for a brief consultation and a fast, customized quotation.
References
- Javdani-Mallak A, Salahshoori I. Environmental pollutants and exosomes: A new paradigm in environmental health and disease. Science of The Total Environment. 2024, 925: 171774.
- Krokidis M G, Pucha K A, Mustapic M, et al. Lipidomic analysis of plasma extracellular vesicles derived from Alzheimer's disease patients. Cells. 2024, 13(8): 702.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.