Oncology Exosome Profiling and Functional Analysis
Tumor-derived exosomes (TDEs) are the architects of the tumor microenvironment. They do not act alone; they orchestrate a complex network of immune suppression, drug resistance, and metastatic niche preparation. For oncology researchers, studying exosomes requires more than a single assay—it requires a multidimensional approach combining deep molecular profiling with rigorous phenotypic validation.
We provide a holistic Oncology Exosome Research Solution. Unlike generic providers, our platform is engineered for the complexity of cancer. Whether you need to identify Liquid Biopsy Biomarkers from plasma, map the Immune Evasion mechanisms mediated by exosomal PD-L1, or track Organotropic Metastasis in vivo, we offer an integrated portfolio of services to accelerate your discovery from bench to clinic.
Critical Research Frontiers in Oncology
Current oncology research is focused on decoding the complex communication networks that drive tumor evolution and treatment failure.
- Tumor Heterogeneity & Plasticity: Tumors are not static; they continuously evolve. Capturing this heterogeneity—specifically the presence of rare, aggressive subclones—through non-invasive liquid biopsy remains a significant challenge.
- The Pre-Metastatic Niche: Before tumor cells spread, they send exosomal signals to prepare distant organs (the "soil"). Understanding how these signals modify the lung, liver, or bone microenvironment is key to preventing metastasis.
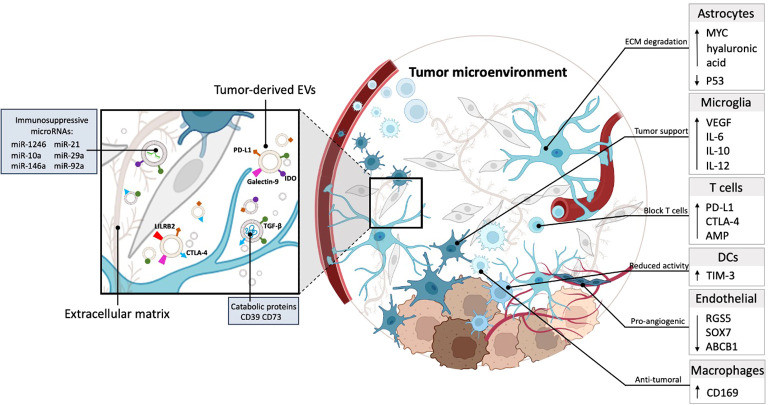
- Immune Evasion Mechanisms: Despite the success of immunotherapy, many tumors remain "cold." Research is intensely focused on how tumor-derived exosomes suppress T-cell activity and reprogram macrophages to support tumor growth.
- Minimal Residual Disease (MRD): Detecting microscopic disease remaining after curative treatment is the frontier of recurrence prevention, requiring ultra-sensitive detection of circulating markers.
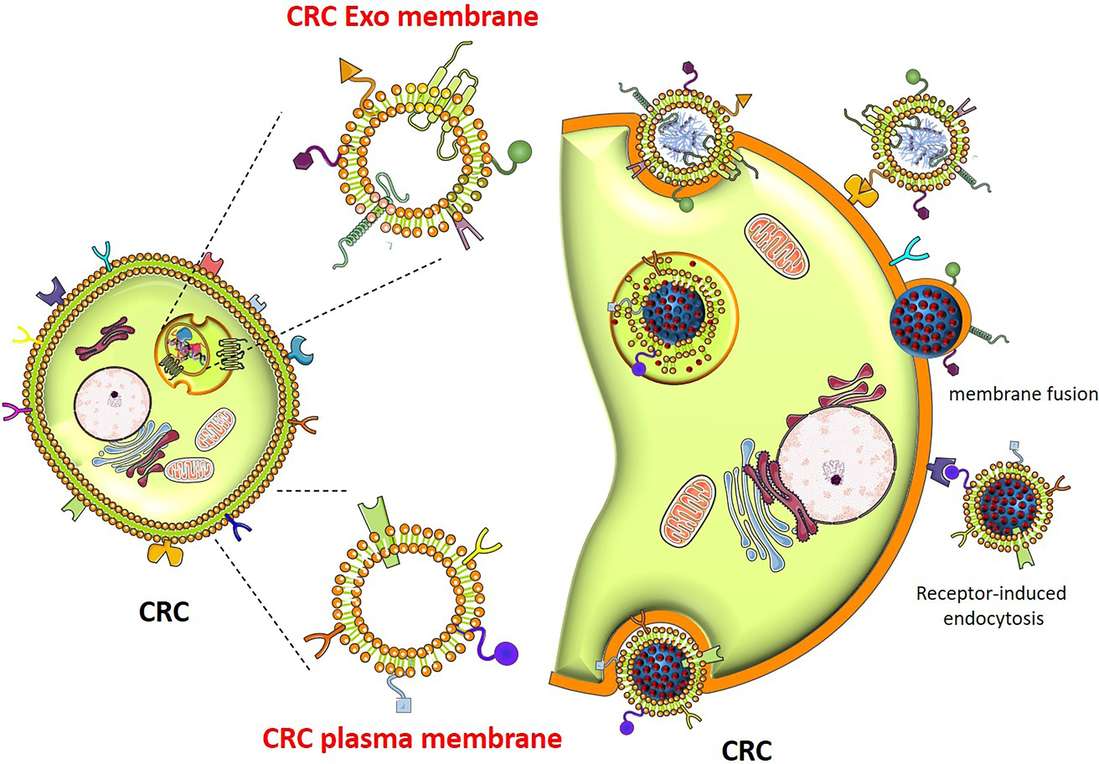
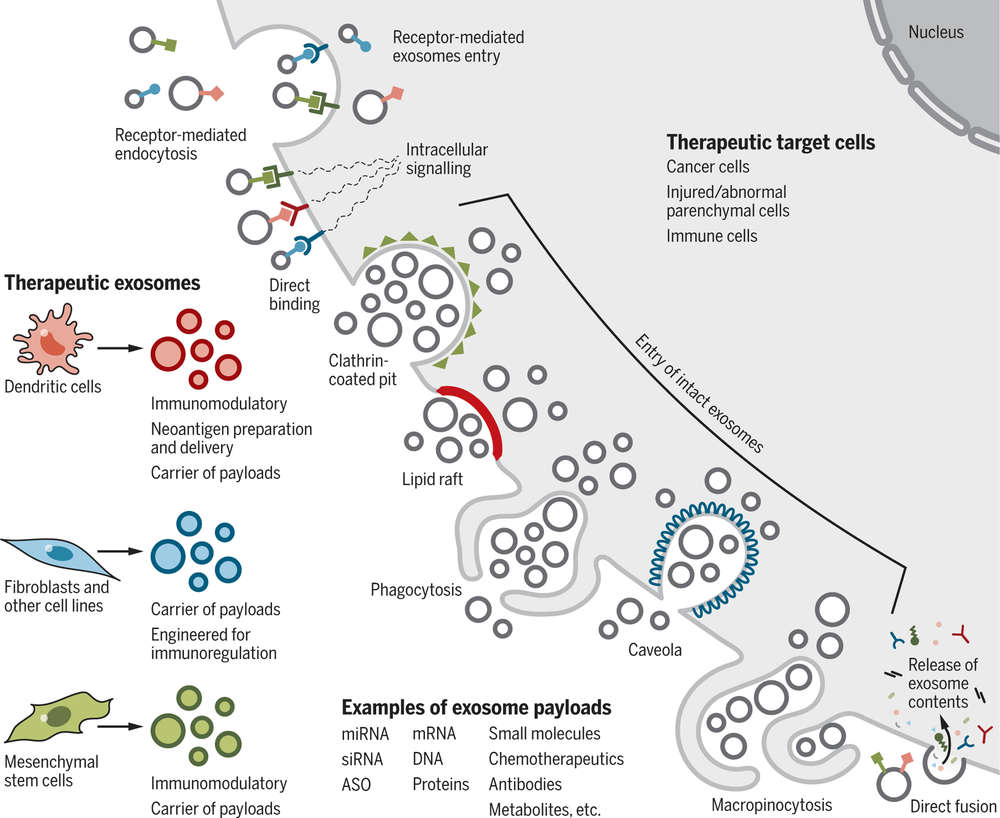 Figure 1. Mechanisms of cellular uptake of therapeutic exosomes and their impact on target cells. (Kalluri R, et al., 2020)
Figure 1. Mechanisms of cellular uptake of therapeutic exosomes and their impact on target cells. (Kalluri R, et al., 2020)
Comprehensive Service Portfolio for Oncology
We offer a discovery-to-function pipeline designed to handle the complexity of tumor samples.
| Research Phase | Our Specialized Approach & Solution | Key Services Applied |
|---|---|---|
| Biomarker Discovery (Diagnosis) | Multi-Omics Profiling: We perform Small RNA-Seq and Label-Free Proteomics on plasma/urine exosomes to identify diagnostic signatures (e.g., miRNAs, specific surface proteins). | Exosome Biomarker Protein Screening, Exosomal Small RNA and miRNA Sequencing |
| TME & Immune Modulation (Mechanism) | Immune Suppression Assays: We co-culture TDEs with T-cells, NK cells, or Macrophages. We measure endpoints like PD-L1 expression, cytokine release, and immune cell proliferation inhibition to validate immune evasion. | Immunomodulation and Inflammation Assays |
| Metastasis & Angiogenesis (Function) | Phenotypic Assays: We utilize Matrigel Invasion Chambers and HUVEC Tube Formation assays to quantify the ability of exosomes to promote cancer cell invasiveness and new blood vessel growth. | Angiogenesis and Stem Cell Functional Assays |
| In Vivo Validation (Animal Models) | Biodistribution & Xenografts: We label TDEs to track their Organotropism (homing to specific organs) in mice. We also perform co-injection studies to assess if exosomes accelerate tumor growth or metastasis in vivo. | Exosome Organoid-based Functional Assays |
Core Technologies for Cancer Biology
We highlight specialized technologies that go beyond standard characterization, specifically designed for oncology applications.
Immune Checkpoint & TME Analysis
Decoding Immune Evasion: Cancer exosomes often carry checkpoint proteins like PD-L1 to deactivate the immune system. Our specialized Immune Modulation Platform allows you to detect exosomal PD-L1 via Flow Cytometry and functionally validate its activity by measuring the suppression of CD8+ T-cell killing capacity in vitro. This is critical for immuno-oncology research.
Organotropic Biodistribution Tracking
Mapping Metastasis: Why do some cancers spread to the lung and others to the liver? We use high-resolution IVIS Fluorescence Imaging to track the fate of injected exosomes. By analyzing the "biodistribution profile," we can determine if your exosomes exhibit Organotropism, homing specifically to the pre-metastatic niche, validating their role in distant spread.
Exosome-Mediated Drug Resistance Models
The "Bystander Effect": We develop specific co-culture models to study acquired resistance. We isolate exosomes from drug-resistant cell lines (e.g., Cisplatin-resistant) and treat sensitive cells. We then measure the shift in IC50 values (drug sensitivity) to prove that resistance traits are horizontally transferred via exosomal cargo.
Application Spotlight: Exosome Integrins Dictate Metastasis
This analysis highlights one of the most significant discoveries in cancer biology: that exosome surface proteins predict where a tumor will metastasize.
Featured Technologies:
- Proteomic Profiling
- Biodistribution Imaging
Literature Interpretation:
Researchers investigated the mechanisms underlying organotropic metastasis to determine why specific cancers preferentially spread to distinct organs, such as the lung or liver. By performing comprehensive proteomic profiling of tumor-derived exosomes, the study revealed that specific Integrin proteins on the exosome surface act as molecular "ZIP codes" for tissue targeting. Specifically, exosomes enriched with Integrin alpha-6-beta-4 were found to home to the lung, while those carrying Integrin alpha-v-beta-5 targeted the liver. Crucially, these exosomes accumulated in the target tissues well before the tumor cells arrived, modifying the local microenvironment to prepare a pre-metastatic niche. This groundbreaking study demonstrates that profiling exosomal surface proteins can accurately predict future metastasis sites, validating the essential role of proteomic analysis in understanding cancer progression.
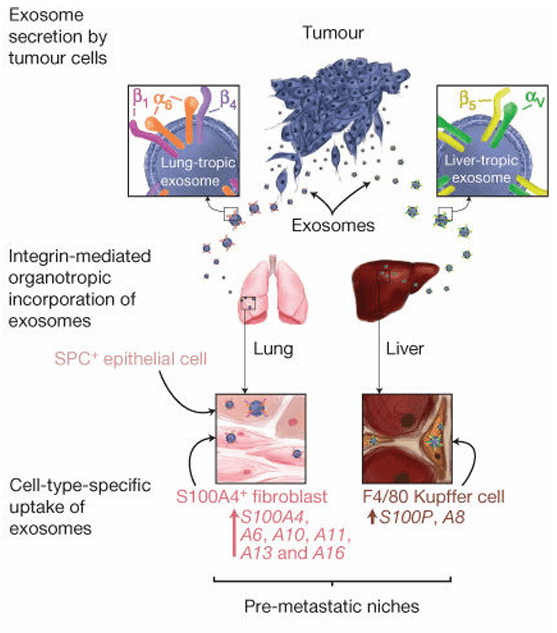 Figure 2. Exosome-mediated tumor dissemination model showing organ-specific uptake by resident cells. (Hoshino A, et al., 2015)
Figure 2. Exosome-mediated tumor dissemination model showing organ-specific uptake by resident cells. (Hoshino A, et al., 2015)
Start Your Oncology Discovery
Leverage our comprehensive platform to accelerate your discovery, from biomarkers to therapeutic targets.
How It Works: Our Project Pathway
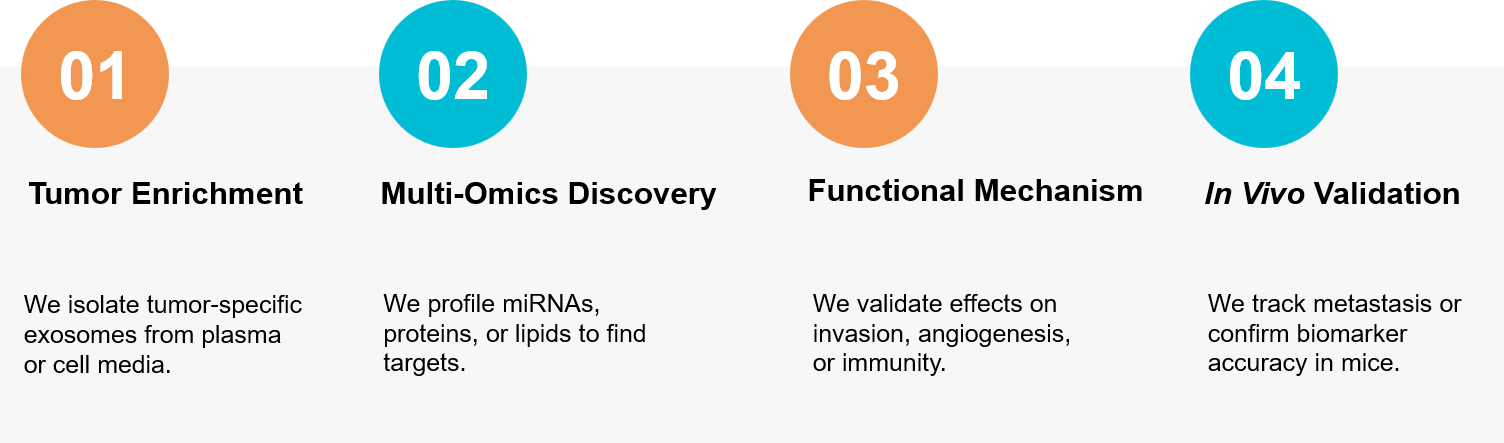 Figure 3. Our comprehensive workflow for profiling tumor-derived exosomes and validating their functional role. (Creative Biostructure)
Figure 3. Our comprehensive workflow for profiling tumor-derived exosomes and validating their functional role. (Creative Biostructure)
Ready to uncover the secrets of the tumor microenvironment? Our oncology experts are available to build a custom study plan tailored to your specific cancer model. Contact us today to discuss your project.
References
- Kalluri R, LeBleu VS. The biology , function , and biomedical applications of exosomes. Science. 2020 Feb 7;367(6478):eaau6977.
- Hoshino A, Costa-Silva B, Shen TL, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015 Nov 19;527(7578):329-35.