Exosome Multi-Omics Integration Services
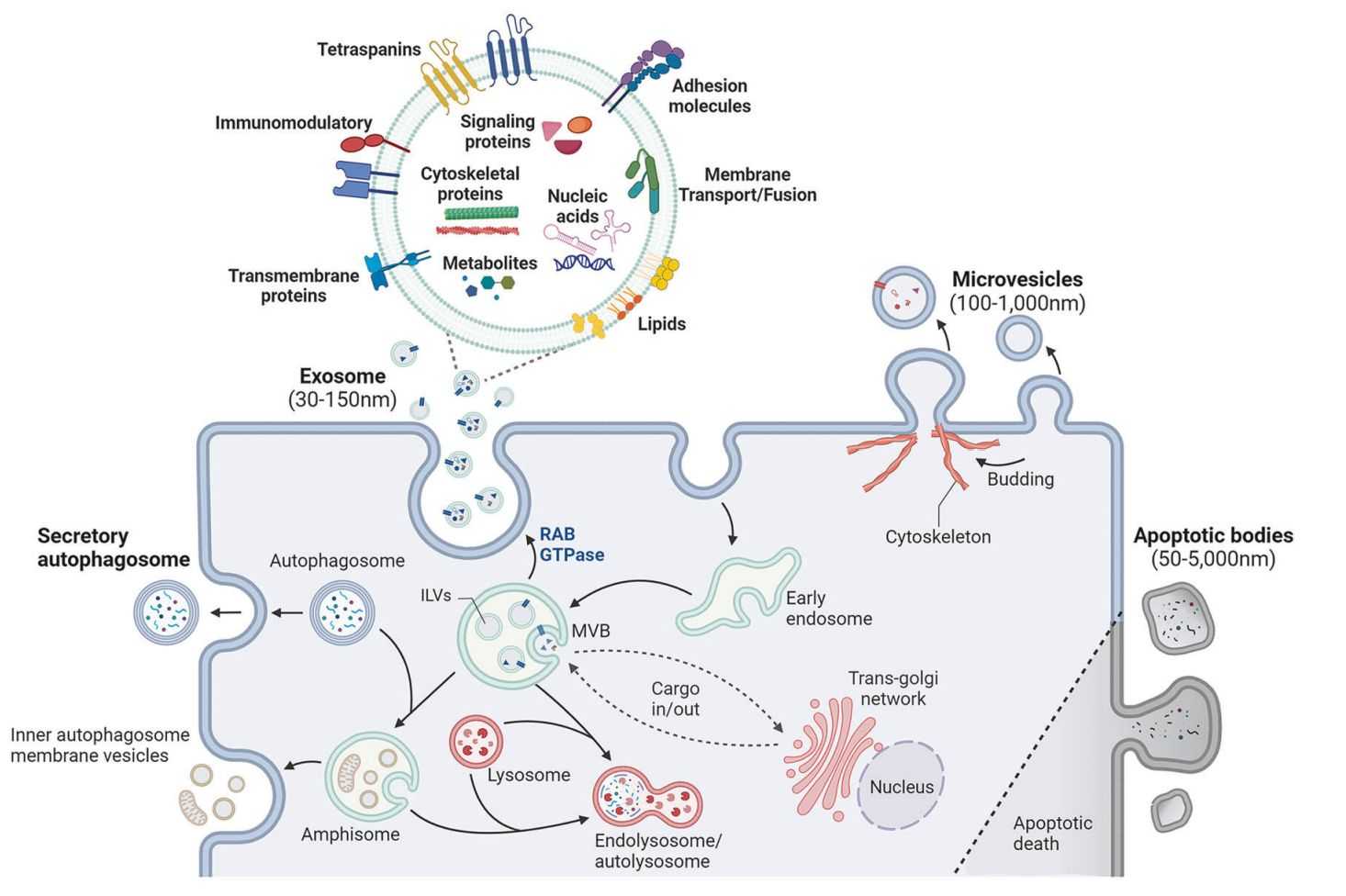
In the rapidly evolving field of extracellular vesicle (EV) research, relying on a single layer of biological data, whether transcriptomic, proteomic, or metabolomic, often yields an incomplete picture. Biological systems are inherently interconnected, as genes regulate proteins, proteins drive metabolic fluxes, and lipids provide the structural context for these interactions.
Creative Biostructure offers comprehensive exosome multi-omics integration services designed to bridge these gaps. By combining high-throughput data acquisition with advanced systems biology bioinformatics, we enable researchers to correlate genotype with phenotype, uncovering the complex regulatory networks hidden within exosomes. Our service transforms fragmented datasets into a unified, actionable biological story, essential for biomarker discovery, mechanism of action (MOA) studies, and therapeutic development.
The Power of Multi-Dimensional Profiling
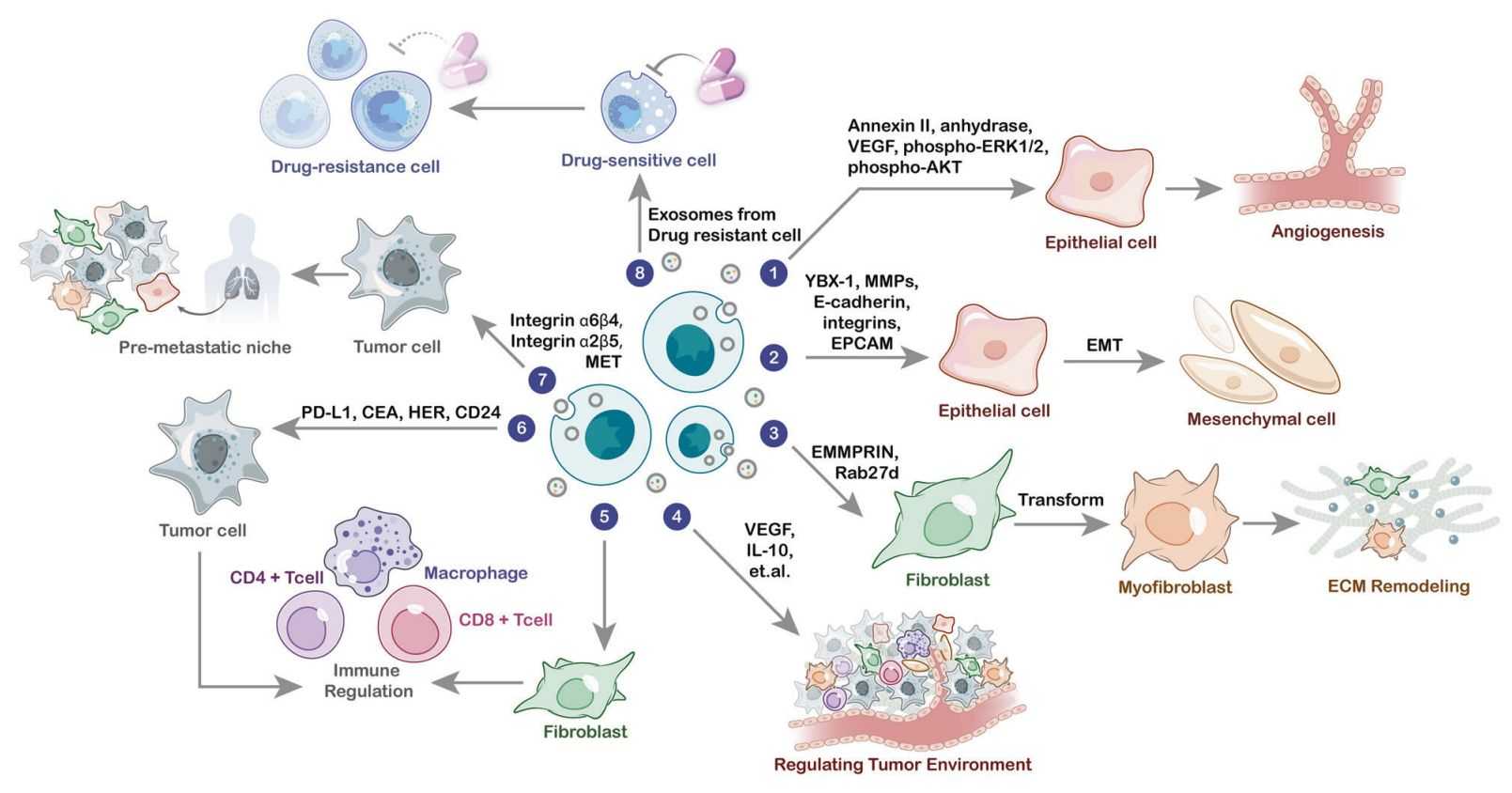
Exosomes are not merely waste bins; they are sophisticated delivery vehicles reflecting the dynamic state of their cell of origin. To fully understand their function, one must analyze them as complete systems.
Our integrated approach addresses the "data silos" problem in EV research. We utilize advanced integration algorithms to merge heterogeneous datasets, allowing you to:
- Validate findings across omics layers: Confirm if increased mRNA levels actually result in higher protein abundance.
- Identify key regulatory hubs: Pinpoint specific transcription factors or enzymes that drive systemic changes.
- Discover composite biomarkers: Develop robust diagnostic panels combining RNAs, proteins, and lipids for higher sensitivity and specificity.
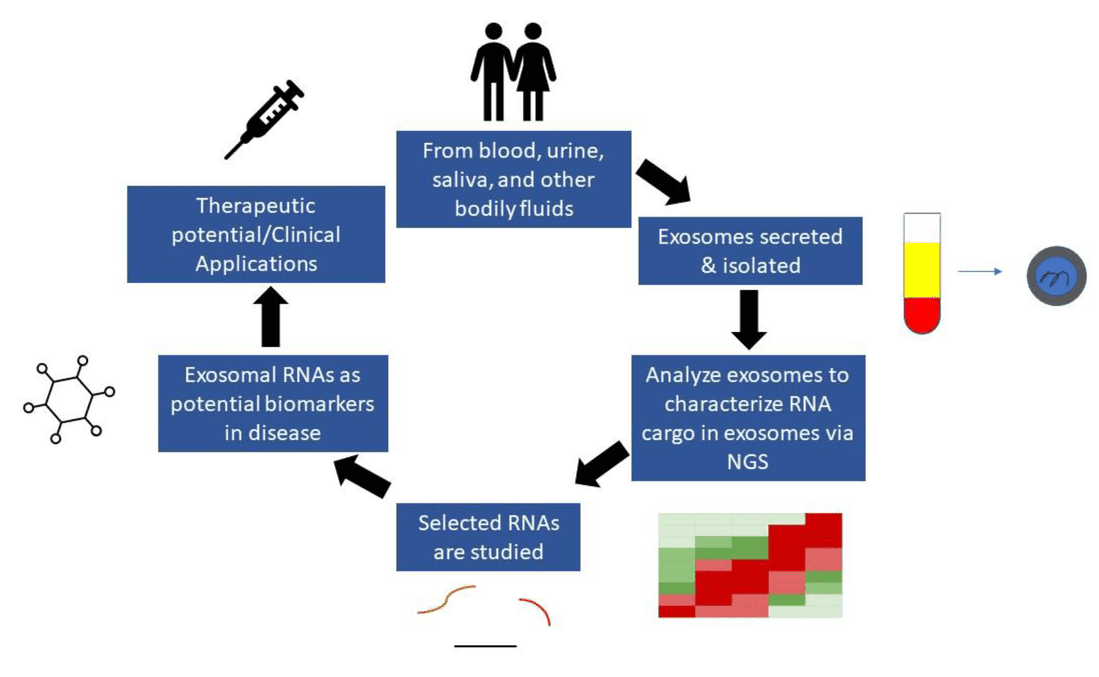
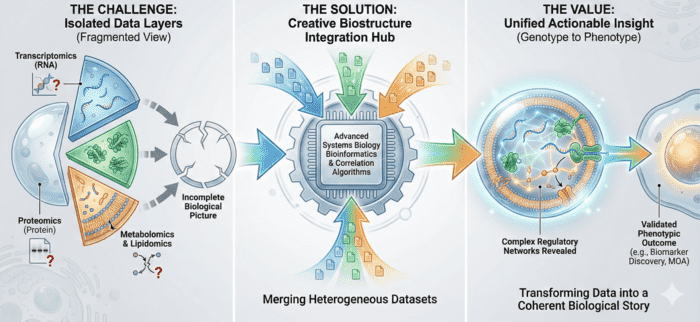 Figure 1. Exosome Multi-Omics Integration for Holistic Biomarker and Mechanism Discovery. (Creative Biostructure)
Figure 1. Exosome Multi-Omics Integration for Holistic Biomarker and Mechanism Discovery. (Creative Biostructure)
Our Core Integration Modules
Creative Biostructure has structured our multi-omics capabilities into two primary pillars, ensuring deep coverage of the exosomal cargo.
Exosome Proteomics and RNA Sequencing Integration
This module focuses on the "Central Dogma" within exosomes. By performing parallel RNA-Seq (mRNA, miRNA, lncRNA, circRNA) and LC-MS/MS-based proteomics on the same sample batch, we reveal post-transcriptional regulation mechanisms.
- Dual-Extraction Protocols: We utilize optimized extraction methods to isolate high-quality RNA and proteins from low-input exosomal samples without cross-contamination.
- Correlation Analysis: We map mRNA-protein pairs to assess concordance, revealing translation efficiency and protein stability.
- miRNA Target Validation: By integrating miRNA abundance with proteomic downregulation, we experimentally validate predicted miRNA targets in a high-throughput manner.
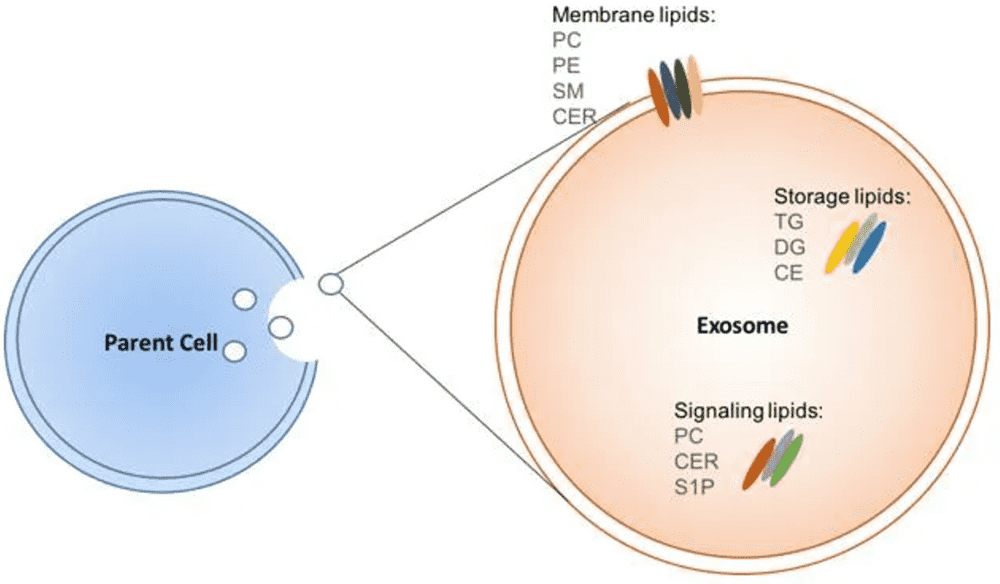
Exosome Lipidomics and Metabolomics Profiling
Exosomes are lipid-bound vesicles that carry metabolic enzymes and intermediates capable of reprogramming recipient cells. This module analyzes the structural composition and the functional metabolic payload.
- Membrane Fluidity & Stability: Lipidomic profiling (phospholipids, sphingolipids, sterols) helps predict exosome stability, uptake efficiency, and tissue tropism.
- Metabolic Reprogramming: Metabolomic analysis reveals how exosomes influence the metabolic state of the tumor microenvironment (TME) or immune system.
- Lipid-Protein Interactome: We analyze how specific lipid species recruit signaling proteins to the exosomal surface.
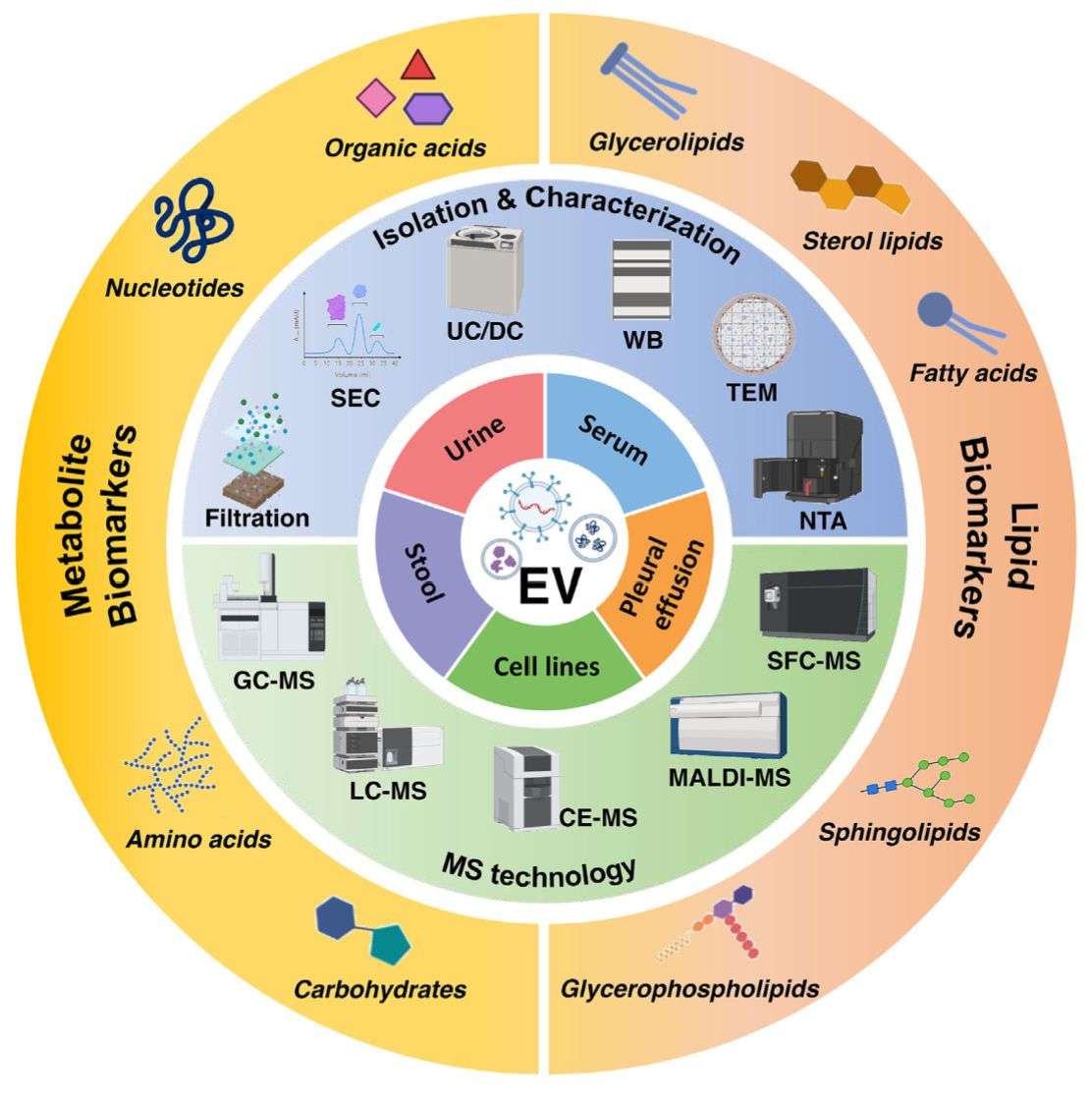
Our Workflow from Sample to Biological Interpretation
Achieving high-quality multi-omics data requires rigorous standardization from the very first step. We adhere to MISEV (Minimal Information for Studies of Extracellular Vesicles) guidelines to ensure reproducibility.
Sample Consultation & Design
We define the optimal sample volume and replicates based on your target sensitivity (e.g., targeted vs. discovery mode).
Standardized Isolation
Using technologies such as Ultracentrifugation or Size Exclusion Chromatography (SEC), we isolate EVs while minimizing non-vesicular contamination (like lipoproteins or albumin) that can skew multi-omics data.
Parallel Cargo Extraction
Our proprietary workflows allow for the simultaneous fractionation of nucleic acids, proteins, and metabolites from limited sample inputs.
High-Throughput Data Acquisition
- Genomics: Illumina NovaSeq or PacBio platforms.
- Mass Spectrometry: Thermo Fisher Orbitrap Exploris and Triple Quadrupole systems for deep coverage of the proteome and metabolome.
Bioinformatic Integration
The raw data enters our computational pipeline for normalization, statistical analysis, and network integration.
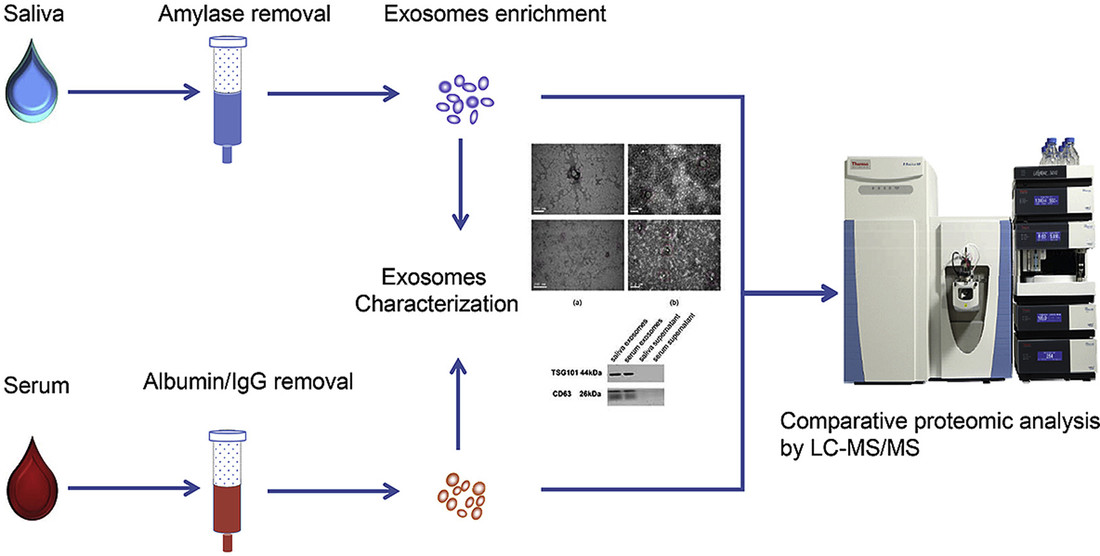
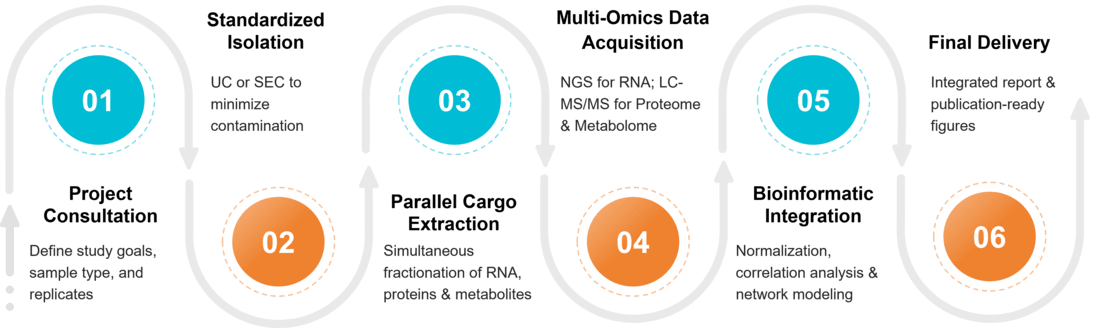 Figure 2. Project Workflow for Exosome Multi-Omics Integration. (Creative Biostructure)
Figure 2. Project Workflow for Exosome Multi-Omics Integration. (Creative Biostructure)
Advanced Bioinformatics
The true value of our service lies in our post-acquisition analysis. Our bioinformaticians employ advanced statistical models to move beyond simple Venn diagrams.
| Analysis Module | Methodologies & Databases | Objective & Biological Insight |
|---|---|---|
| Dimensionality Reduction | Principal Component Analysis (PCA), t-SNE | Global Pattern Visualization: Reduces data complexity to visualize sample clustering and detect outliers across combined multi-omics datasets. |
| Pathway Co-Enrichment | KEGG, Reactome, Gene Ontology (GO) | Functional Convergence: Identifies biological pathways that are significantly enriched simultaneously across the transcriptome, proteome, and metabolome. |
| Interaction Network Construction | PPI Networks, Metabolic Flux Analysis | Systemic Connectivity: Constructs Protein-Protein Interaction networks overlaid with metabolic flux data to map the functional connectivity of exosomal cargo. |
| Canonical Correlation Analysis (CCA) | Multivariate Statistical Analysis | Cross-Omics Correlation: Identifies linear combinations of variables that exhibit the highest correlation between two distinct datasets (e.g., linking specific lipids to protein expression). |
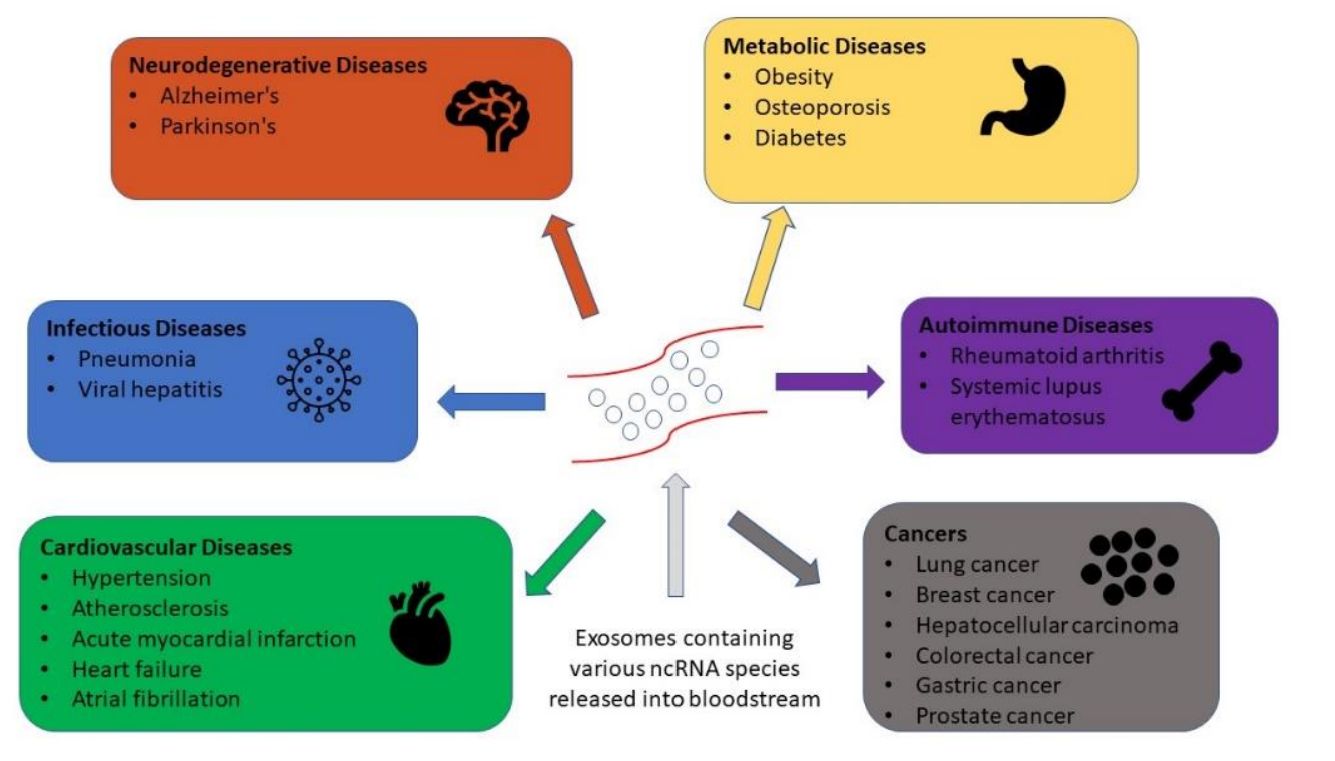
Key Research Areas Supported by Exosome Multi-Omics Integration
- Oncology & Liquid Biopsy: Distinguishing tumor-derived exosomes from host exosomes by identifying specific mutation signatures (RNA) alongside surface markers (Protein) and metabolic shifts.
- Neuroscience: Investigating exosomes crossing the Blood-Brain Barrier (BBB) to identify composite biomarkers for Alzheimer's and Parkinson's disease.
- Immunology: Understanding how immune cell-derived exosomes modulate inflammation via lipid mediators and cytokine transport.
- Regenerative Medicine: Characterizing the "potency" of MSC-derived exosomes by correlating their protein content with their lipid membrane stability.
Why Choose Creative Biostructure
- True "Omics" Expertise: We are not just a sequencing shop or a mass spec lab. We are integrated biology experts with years of experience handling the unique challenges of low-abundance exosomal samples.
- Customizable Pipelines: Whether you need a broad discovery approach (untargeted) or precise quantification of specific molecules (targeted MRM), we tailor the pipeline to your hypothesis.
- Publication-Ready Output: We deliver comprehensive reports containing high-resolution visualizations (circos plots, heatmaps, interaction networks) ready for top-tier journal submission.
- Technical Transparency: We provide detailed method descriptions, including QC metrics for isolation purity and sequencing depth, ensuring you have full confidence in your data.
Case Study
Case: Multi-Omics Integration of Circulating Exosomes in OSA
Background
This study profiled plasma exosomes from Obstructive Sleep Apnea (OSA) patients before and after treatment to bridge the gap between molecular cargo and clinical phenotypes.
Methods
A Multi-Block Discriminant Analysis (DIABLO) framework was applied to integrate heterogeneous datasets, including lipids, proteins, miRNAs, and clinical features. Circos plots were generated to visualize strong cross-layer correlations.
Results
The analysis pinpointed key discriminators located in the "outer circle" of the variable plot, indicating their high contribution to group separation. A dense regulatory network was revealed, linking specific exosomal signatures directly to clinical improvements, such as AHI reduction and systolic blood pressure changes.
Conclusion
This integrative workflow successfully transformed fragmented omics data into a unified biological narrative. By correlating multi-dimensional cargo with phenotypic data, the study identified robust composite biomarkers that would remain invisible in single-omics analysis.
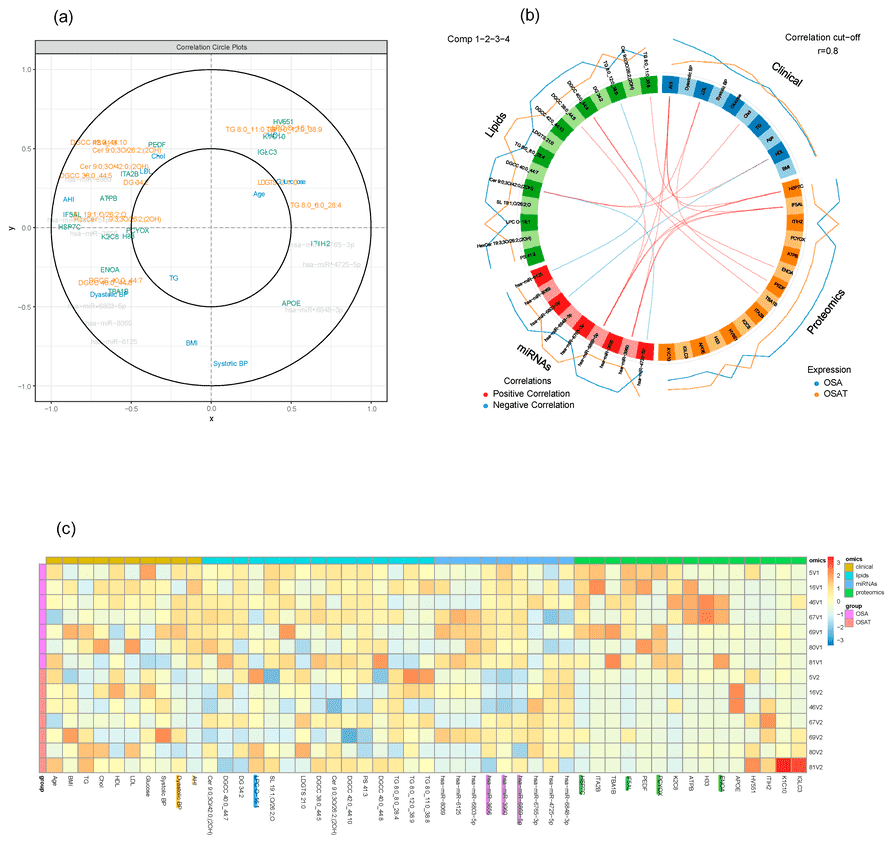 Figure 3. Multi-block analysis integrating data and exosomal cargos (lipids, proteins, miRNAs) from OSA and OSAT samples. (Khalyfa A, et al., 2023)
Figure 3. Multi-block analysis integrating data and exosomal cargos (lipids, proteins, miRNAs) from OSA and OSAT samples. (Khalyfa A, et al., 2023)
Ready to bridge the gap between genotype and phenotype in exosome research? Whether your goal is composite biomarker discovery, regulatory network mapping, or mechanism of action validation, Creative Biostructure delivers advanced multi-omics integration to reveal the complete biological picture. We transform fragmented datasets into actionable insights tailored to your specific study. Contact us for a brief consultation and a fast, customized quotation.
Reference
- Khalyfa A, Marin J M, Sanz-Rubio D, et al. Multi-omics analysis of circulating exosomes in adherent long-term treated OSA patients. International Journal of Molecular Sciences. 2023, 24(22): 16074.
Frequently Asked Questions
For any inquiries, our support team is ready to help you get technical support for your research and maximize your experience with Creative Biostructure.